













| General Information: |
|
| Name(s) found: |
ape2 /
SPBC1921.05
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 882 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
proteolysis
[ISS cellular catabolic process [NAS] |
| Molecular Function: |
zinc ion binding
[IEA]
aminopeptidase activity [ISS metallopeptidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQVTIPKSTS KEDDKNRNLL PKNVKPIHYD LSLYPDLETF TYGGKVVVTL DVLEDSNSIT 60
61 LHGINLRILT AALEWGSQTV WASEVSYGDE RIVLQFPSTV PANSVAVLTL PFTARISSGM 120
121 EGFYRSSYVD SDGNTKYLAT TQMEPTSARR AFPCWDEPAL KATFTIDITA KENYTILSNM 180
181 NAVEETVKDG LKTARFAETC RMSTYLLAWI VAELEYVEYF TPGKHCPRLP VRVYTTPGFS 240
241 EQGKFAAELG AKTLDFFSGV FGEPYPLPKC DMVAIPDFEA GAMENWGLVT YRLAAILVSE 300
301 DSAATVIERV AEVVQHELAH QWFGNLVTMQ FWDGLWLNEG FATWMSWFSC NHFYPEWKVW 360
361 ESYVTDNLQS ALSLDALRSS HPIEVPIMHD YEINQIFDAI SYSKGSCVIR MVSKYVGEDT 420
421 FIKGIQKYIS KHRYGNTVTE DLWAALSAES GQDISSTMHN WTKKTGYPVL SVSETNDGEL 480
481 LIEQHRFLST GDVKPEEDTV IYWAPLKLKT MKDGKAVVDE KAVLSDRSKK IKVDKEALES 540
541 YKLNSEQSGI YRVNYSADHL KKLSQIAVEK PDYLSVEDRA GLIADVASLS RAGYGKVSST 600
601 LDLIKTWKDE PNFVVFAEML ARLNGIKSTL RFESSDIIAA MKKLVLEVSA TKAHSLGWEF 660
661 KANDDHIIRQ FKSTVYNYAG LFGDDKVVKD ALSKFDAYAS GNKSAINDNL RSAVFNIAIR 720
721 YGGAKSWDQL LEIYTKTNDP YVRNSCLRAF GVTEDEKYIQ KTLDLTLDPI VKEQDIYLIL 780
781 VTLSTHKNGV LAMWKFATSN WDKLLSRLPV AGTMRGYVVR FVTSGFTHAS AIDKIKEFFA 840
841 DKDTKLYERA LQQSLDTISA NSSFIDKSLD DITRWLKENR YM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
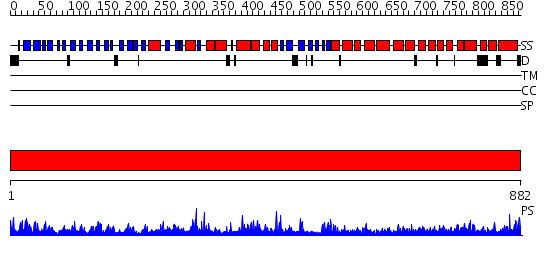
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..882] | 156.0 | Crystal structures of the tricorn interacting facor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)