













| General Information: |
|
| Name(s) found: |
ppk26 /
SPBC336.14c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA PAN complex [ISS cytoplasm [ISS cytosol [IDA |
| Biological Process: |
signal transduction
[RCA mRNA 3'-end processing [ISS protein amino acid phosphorylation [IEA] postreplication repair [ISS |
| Molecular Function: |
ATP binding
[IEA]
zinc ion binding [IEA] protein serine/threonine kinase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVRKNSPAS PKPTSRSRES SRSPSVTDLK DHSKAKRTLC RNILLYGSCK HSENGCAFRH 60
61 DGPFIPSSEN LEQYSVKKKL NAASASFQPV RALPVKAAGA AVFVPKSQEK SLFLSRERTP 120
121 VALSPGSHAI NPYNNASMLV NEPFHDDSGV YYTRQNQFKP TLYNLYNPQP SPMPTLLPYE 180
181 RTVNGLFIDD TTRERIERKS EASRQTISAL PSIISSYTSL APLNTKLYRY KEQIGFSSWT 240
241 YKCTSSIDGN AYVLKRLQDC SINIDTSTVD KLKNVFHPNI VPFHSAFHTD TFHDSSLLLI 300
301 YDFYPCTTTL GELYLNNSKN SVKLEENRKI PERELWNYFF QLTIALSYLH KSGFACNKLT 360
361 PSRILVDQTE RIRISGCADY ELVVSNKPPL EERKKQDFVD LGVVIANLAT GRTDMDMSSA 420
421 ARAIYSTYSR EFYKAVLYFV SEVPEDKNLE LFLQNHIESF FPIMSSPYVE CEKMERKISD 480
481 AFQHGRFFNI LCKIMFIIDN NRASREYPIA REKEISLIYL LRDYLFHQID EDECPVIDLY 540
541 QVLNRLGKLD AGINQAIALI SRDELDCVSV SYGELKAWLD NVYEMEINS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
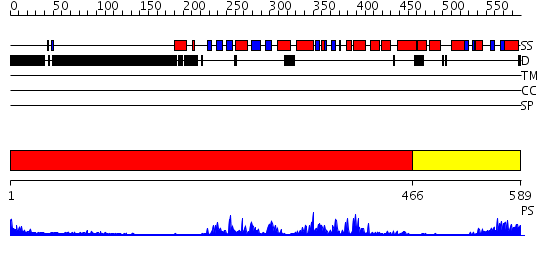
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..465] | 44.154902 | No description for 2h8hA was found. | |
| 2 | View Details | [466..589] | 36.69897 | No description for 2acxA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)