













| General Information: |
|
| Name(s) found: |
raf2 /
SPCC970.07c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 636 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mating-type region heterochromatin
[TAS nucleus [IDA cytoplasm [IDA CLRC ubiquitin ligase complex [IDA nuclear centromeric heterochromatin [IC |
| Biological Process: |
positive regulation of histone H3-K9 methylation
[IMP chromatin silencing at centromere [IMP negative regulation of histone H3-K4 methylation [IMP regulation of mating type switching [IMP meiotic chromosome segregation [IMP mitotic sister chromatid segregation [IMP chromatin silencing at silent mating-type cassette [IMP chromatin silencing at telomere [IMP meiotic sister chromatid cohesion, centromeric [IMP cellular protein localization [IGI chromatin silencing by small RNA [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPVRAEKKR KTDLIEQVCV TNKAGELVDL EDVLEYGPYS LTGILSSEKD EQEPLFDESI 60
61 VLGYSIKISP VWTYNLKDVP EKETMSIWIV TPQRRYGILS PSSEYKAIYE QISEKNRLFY 120
121 LIKTKFKDDM ISGTLEDYDN YIEVLKEKLE LPSCFQAILL VQKHIRFLLT QMVATSSLHV 180
181 WSESPFFIRI RSSYEHLILQ INKNIYNARQ ERKKSKLSSN NPSDNNTTMK SSLNQALTLI 240
241 NLPEQPFSIS SPTATPQLGV VKRTSPLRFP LNDIWLSGLR IVDPNIESIS LWKRIQVSTS 300
301 PKHQRYISLQ EVCSVIAQQL QITNLEALNK LSSHGETLLQ IMHTAFTWRG TKLFNDIKHA 360
361 IGFRSSVQQA RSQFRGYCYD YLFMHCNNGE KTSLHLLRTL ICMKLDFSNA QLAAKILFHF 420
421 LLFDIGSGLS GSDYTYEQYI NHSAVAFSFT EEIFEKNFVT VLPDFVKLFS ISFGYWPAFS 480
481 FYDELLKLLR NKYPKVYSST PNLCDQVWLD RTNLFPCNRS TRSTLPYRPT KLLDLASASS 540
541 CLSKKETDFK QDTGLYSYNL EKVEALKVSP DLQTGIWSCP VQNCLYFAVC DNPYKPSQVI 600
601 YDHLLGHVDS KFIFKTPSNS VRSFTNKLEH IMYNIN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
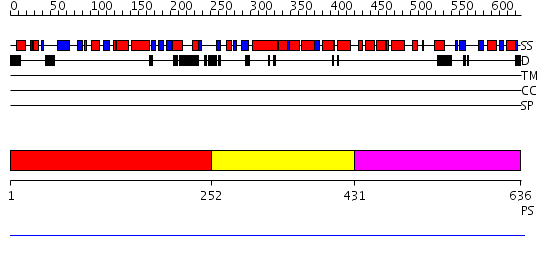
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..251] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [252..430] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [431..636] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)