| Desc: |
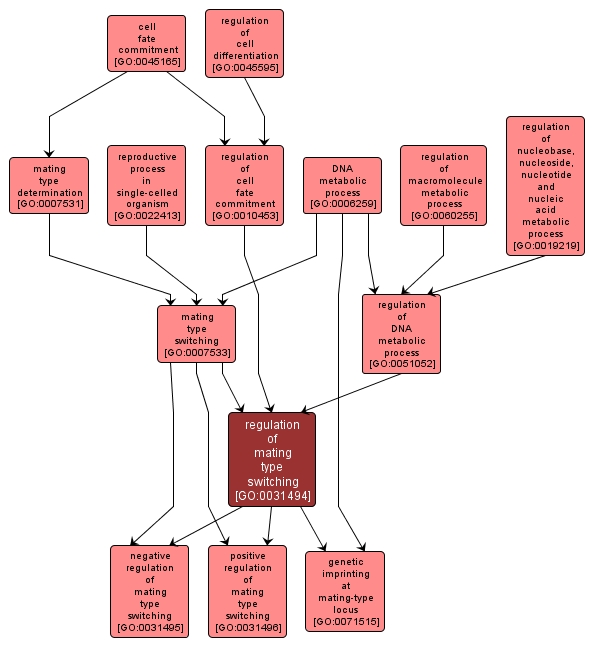
Any process that modulates the frequency, rate or extent of mating type switching, the conversion of a single-cell organism from one mating type to another by the precise replacement of a DNA sequence at the expressed mating type locus with a copy of a sequence from a donor locus. |