













| General Information: |
|
| Name(s) found: |
ubx2 /
SPAC2C4.15c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 427 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS cytoplasm [ISS |
| Biological Process: |
proteasomal ubiquitin-dependent protein catabolic process
[IGI cellular response to stress [IEP ubiquitin-dependent protein catabolic process [ISS |
| Molecular Function: |
protein binding
[IPI ubiquitin binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGDEASLVA NFCAITNSTP EKAQEYLSVA DGDLSTAITL FFESGGVTDV QSSYIEAPSQ 60
61 TEPVEEIRAP IAPTREVLVD PLADMSAGTS IMGNNFGFGG FPRMNRRQRR RMGIFDQSPS 120
121 QIPFPSSNTE DSSEESDSSS RASRLAKLFR PPYDIISNLS LDEARIEASS QKRWILVNLQ 180
181 TSTSFECQVL NRDLWKDESV KEVIRAHFLF LQLLDDEEPG MEFKRFYPVR STPHIAILDP 240
241 RTGERVKEWS KSFTPADFVI ALNDFLEGCT LDETSGRKNP LGAKSQKPVE AMSEDEQMHK 300
301 AIAASLGNGN STTESQGESS SQQAESHGVA DDTVHKIDSA ECDAEEPSPG PNVTRIQIRM 360
361 PNGARFIRRF SLTDPVSKVY AYVKGVAEGA DKQPFSLTFQ RKSLWTSLDS TIKEAGIQNT 420
421 ALQFEFQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
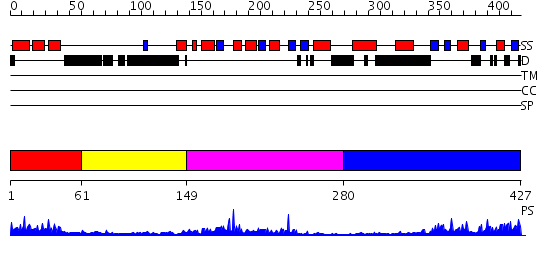
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..60] | 6.0 | No description for 2damA was found. | |
| 2 | View Details | [61..148] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [149..279] | 17.045757 | No description for 2dlxA was found. | |
| 4 | View Details | [280..427] | 17.0 | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)