













| General Information: |
|
| Name(s) found: |
taf4 /
SPAC23G3.09
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 365 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA transcription factor TFIID complex [ISS chromatin [RCA |
| Biological Process: |
RNA polymerase II transcriptional preinitiation complex assembly
[ISS]
positive regulation of transcription [IC regulation of transcription [IC transcription initiation from RNA polymerase II promoter [ISS |
| Molecular Function: |
transcription initiation factor activity
[ISS RNA polymerase II transcription factor activity [ISS general RNA polymerase II transcription factor activity [IC] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLNNPPSKR FGDDDDIPAN KRPKNEEYPE IPGYLGNRKP SYTQNPNSGP GKLHYASPRP 60
61 NNVSSSVGVA SGGDRQEADQ LQDALISCGI QLKEEELNLS TSFYDPSSLN TFALTTEDRS 120
121 RKSDFLNSFV LMQTVSNIVN LHRLKSMDSD IHALISMAVR DYLANLLQKM IVESHHRTSQ 180
181 LHTDNYKQVD NVRQTLANFA YKEYESEERR RTVLNIRRAE HEARLAELNS ASTNEEGSSR 240
241 RRKEQSSSAA AKNISEDAQN RMTNATASIM AGSALPSGGK KYSWMATDMT PMTPAVGGGF 300
301 GIRKKDSNSL KPSSRDGVLP LQQEEKGIIT IRDALAVLEM DREGAGRIFG RGAKAMMRAY 360
361 IRLKD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
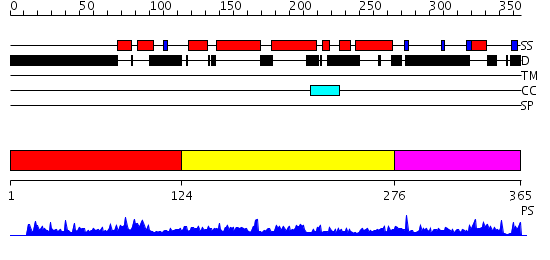
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [124..275] | 19.221849 | TAF(II)-135, (TAF(II)-130, hTAF4), histone fold domain | |
| 3 | View Details | [276..365] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)