













| General Information: |
|
| Name(s) found: |
spt16 /
SPBP8B7.19
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1019 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA FACT complex [IDA nuclear chromatin [ISS spindle pole body [IDA |
| Biological Process: |
DNA repair
[TAS chromatin remodeling [ISS DNA-dependent DNA replication [TAS histone exchange [IC nucleosome assembly [ISS RNA elongation from RNA polymerase II promoter [ISS |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEYEIDEIT FHKRLGILLT SWKNEEDGKT LFQDCDSILV TVGAHDDTNP YQKSTALHTW 60
61 LLGYEFPSTL ILLEKHRITI LTSVNKANML TKLAETKGAA ADVNILKRTK DAEENKKLFE 120
121 KIIEYIRATN KKVGVFPKDK TQGKFINEWD SIFEPVKSEF NLVDASLGLA KCLAIKDEQE 180
181 LANIKGASRV SVAVMSKYFV DELSTYIDQG KKITHSKFSD QMESLIDNEA FFQTKSLKLG 240
241 DIDLDQLEWC YTPIIQSGGS YDLKPSAITD DRNLHGDVVL CSLGFRYKSY CSNVGRTYLF 300
301 DPDSEQQKNY SFLVALQKKL FEYCRDGAVI GDIYTKILGL IRAKRPDLEP NFVRNLGAGI 360
361 GIEFRESSLL VNAKNPRVLQ AGMTLNLSIG FGNLINPHPK NSQSKEYALL LIDTIQITRS 420
421 DPIVFTDSPK AQGDISYFFG EDDSSLEDGV KPRKPPTRGT ATISSHKGKT RSETRDLDDS 480
481 AEKRRVEHQK QLASRKQAEG LQRFAQGSVP SSGIEKPTVK RFESYKRDSQ LPQAIGELRI 540
541 LVDYRAQSII LPIFGRPVPF HISTLKNASK NDEGNFVYLR LNFVSPGQIG GKKDELPFED 600
601 PNAQFIRSFT FRSSNNSRMS QVFKDIQDMK KAATKRETER KEFADVIEQD KLIEIKNKRP 660
661 AHINDVYVRP AIDGKRLPGF IEIHQNGIRY QSPLRSDSHI DLLFSNMKHL FFQPCEGELI 720
721 VLIHVHLKAP IMVGKRKTQD VQFYREVSDI QFDETGNKKR KYMYGDEDEL EQEQEERRRR 780
781 AQLDREFKSF AEKIAEASEG RIELDIPFRE LAFNGVPFRS NVLLQPTTDC LVQLTDTPFT 840
841 VITLNEIEIA HLERVQFGLK NFDLVFIFQD FRRPPIHINT IPMEQLDNVK EWLDSCDICF 900
901 YEGPLNLNWT TIMKTVNEDP IAFFEEGGWG FLGAPSDDEG DDSVEEVSEY EASDADPSDE 960
961 EEEESEEYSE DASEEDGYSE SEVEDEESGE DWDELERKAR QEDAKHDAFE ERPSKKRHR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
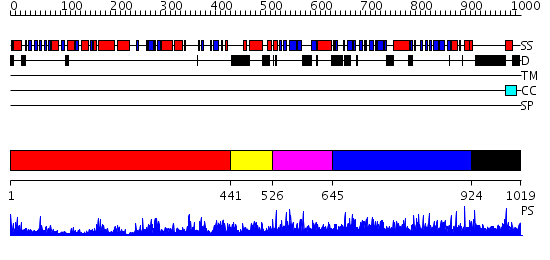
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..440] | 80.69897 | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | |
| 2 | View Details | [441..525] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [526..644] | 36.39794 | No description for PF08644.2 was found. No confident structure predictions are available. | |
| 4 | View Details | [645..923] | 0.965 | No description for 2gcjA was found. | |
| 5 | View Details | [924..1019] | 2.047996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)