













| General Information: |
|
| Name(s) found: |
taf73 /
SPBC15D4.14
[Sanger Pombe]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transcription factor TFIID complex
[TAS cytosol [IDA |
| Biological Process: |
regulation of mitotic metaphase/anaphase transition
[IGI positive regulation of transcription [IC transcription initiation from RNA polymerase II promoter [TAS |
| Molecular Function: |
transcription initiation factor activity
[IC |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQSSYSSGN STSDLNKIVL DYLFKKGYSR TEAMLRLEIS SVGAIPEEQQ DWHIGASETY 60
61 VQVYILLRDW IDGTLDLYKP KLQKVLYPIF VHSYLDLLQK NDPEMAIYFF ESFRIEHEVL 120
121 HGYDIRALAQ LKIASDVEEI EIAQLYRKNK YRINFTRSTF DLLVQFLFEN EVNGSGIIIR 180
181 LLNQYIDIKI TMPQVEGKED PNVQAVYEMD EAKNEQTIIS EQEGIPGHSE QLLDYNKQSI 240
241 QLGLRPLSEE VVTTLKNLLE VKDKDVEGRN EALNKILHPA KNLVELLTEK ENLINESTLE 300
301 SPSDPPLPPT KMKDIDYFVS LLESEQNSLI LGKNATDPSV FMYTMHNTLS AVNCAAFSDD 360
361 ASMFALGCAD SSIHLYSSTN NGPQPLVGSQ NEPLQKSSLI GHTRPVFGVS ISPQKEFILS 420
421 CSEDGFTRLW SKDTKSTIVK YAGHNAPIWD VQFSPFGYYF ATASHDQTAR LWDVEHAAPL 480
481 RVFVGHQNDV DCVSFHPNAA YLATGSSDHT TRMWDVRTGG TVRVFNAHHS PVSALCMSAD 540
541 GLSLASADES GIIKVWDLRS SNQHVSFVKH SSIVYSLSFS YDNKILVSGG ADTDVNFWDL 600
601 TYRNNLSDGE QSPLLFNFKT KNTIIQNLSF TRRNYCLALS SS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
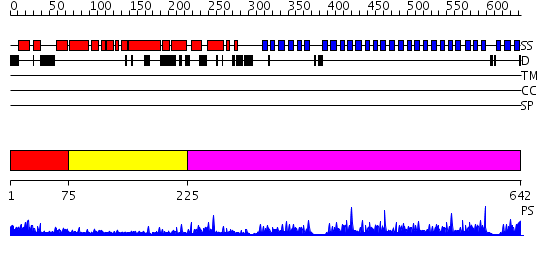
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [75..224] | 18.221849 | No description for 2j49A was found. | |
| 3 | View Details | [225..642] | 74.0 | PAF-AH Holoenzyme: Lis1/Alfa2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)