













| General Information: |
|
| Name(s) found: |
lub1 /
SPBC887.04c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 713 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA protein complex [RCA |
| Biological Process: |
DNA repair
[ISS proteasomal ubiquitin-dependent protein catabolic process [IMP cellular response to stress [IMP histone ubiquitination [NAS |
| Molecular Function: |
protein binding
[IPI ubiquitin binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSYELSREL GGHKQDVRGV CSISNELIGS ASRDGTYSVW EQINGEWTPH FYENHEGFVN 60
61 CVCYVPAIDK NSRGGQDKCG ILQEVGTNSP SYYLFGHESN ICSASALNSE TIITGSWDST 120
121 ARVWALGQCK YVLKGHQSSV WAVLALGEDI FITGSADKLI KIWNGEKLVK SILAHNDCVR 180
181 SLCQIPGGFA SCSNDGVIKL WTSDGEFLYE LHGHTSFVYS LTYIHNQQLI ASCGEDRTIR 240
241 IWKGKECLQC ITLPTTSVWS VSSLPNGDLV CGSSDGFVRI FTVDKVRVAP TEVLKNFEER 300
301 VSQFAISSQE VGDIKKGSLP GLEILSKPGK ADGDVVMVRV NNDVEAYQWS QKENEWKKIG 360
361 QVVDAVGNNR KQLFEGKEYD YVFDVDVADG QAPLKLPYNA TENPYQAANR FLELNQLPLS 420
421 YTDEVVKFIE KNTQGHSLES KKEPNLESQS SNKIKTTIFP VSQLLFSNAN VPAMCQRLRS 480
481 LNNTKSNPLP AKSIDSLERA LSSKKITDTE KNELLETCLS ILDSWSLAER FPALDALRLL 540
541 AINSSSDLAP IFLEVFSRVV KSVPSSGNFE SINVMLALRG LSNVVPNITD AEGVSKLMDC 600
601 LTSTVPQASS AKDFKIAFAT LAMNLSILLI QLNLENTGIE LLSILFSFLD DPSPDNEAFY 660
661 RALMALGTLC TVPDIALAAS QIYHAQSIVH GIAERFSQEM RFVDAEKQIL SLF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
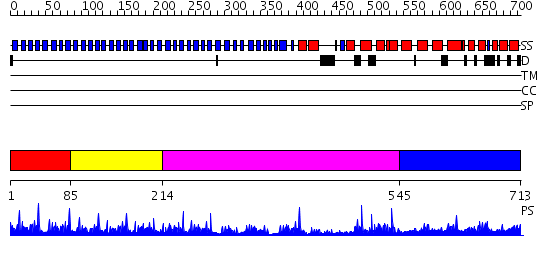
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 64.522879 | No description for 2ovpB was found. | |
| 2 | View Details | [85..213] | 4.154902 | Surface layer protein | |
| 3 | View Details | [214..544] | 11.69897 | Crystal Structure analysis of the beta-propeller protein Ski8p | |
| 4 | View Details | [545..713] | 6.585027 | No description for PF08324.2 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)