













| General Information: |
|
| Name(s) found: |
apn2 /
SPBC3D6.10
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 523 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
base-excision repair
[IC cellular response to reactive oxygen species [IMP |
| Molecular Function: |
DNA binding
[IEA]
phosphodiesterase I activity [ISS endonuclease activity [IEA] double-stranded DNA specific 3'-5' exodeoxyribonuclease activity [IMP DNA-(apurinic or apyrimidinic site) lyase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRILSWNVNG IQNPFNYFPW NKKNSYKEIF QELQADVICV QELKMQKDSF PQQYAVVEGF 60
61 DSYFTFPKIR KGYSGVGFYV KKDVAIPVKA EEGITGILPV RGQKYSYSEA PEHEKIGFFP 120
121 KDIDRKTANW IDSEGRCILL DFQMFILIGV YCPVNSGENR LEYRRAFYKA LRERIERLIK 180
181 EGNRKIILVG DVNILCNPID TADQKDIIRE SLIPSIMESR QWIRDLLLPS RLGLLLDIGR 240
241 IQHPTRKGMF TCWNTRLNTR PTNYGTRIDY TLATPDLLPW VQDADIMAEV MGSDHCPVYL 300
301 DLKEEYEGKK LSNFLSHSKE PPLLSTAHHS AYRPSKNIHS MFQHFNSMKK NKNNSPTQSE 360
361 NVSASASSGS SPTVSRANSV IDVDAYPPEK RRRKEQSKLL SFFAKQKEEK EETNKTEDVS 420
421 IEVLDNNNES DIGLTVKKKV ENGNAWKQIF SERAPPLCEG HKEPCKYLTV RKPGINYGRK 480
481 FWICARPVGE LIKNSNAVSE EDTQPFQCRF FIWDSDWRAN SKD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
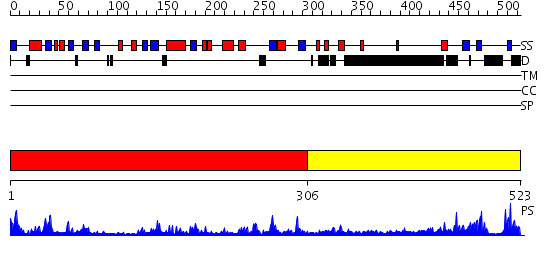
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..305] | 54.69897 | DNA repair endonuclease Hap1 | |
| 2 | View Details | [306..523] | 11.172993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)