













| General Information: |
|
| Name(s) found: |
mbx2 /
SPBC317.01
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 372 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA nucleus [IDA cytosol [IDA |
| Biological Process: |
fungal-type cell wall biogenesis
[IMP regulation of transcription, DNA-dependent [IEA] 4,6-pyruvylated galactose residue biosynthetic process [IMP gene-specific transcription from RNA polymerase II promoter [RCA |
| Molecular Function: |
transcription factor activity
[IEA]
specific RNA polymerase II transcription factor activity [RCA sequence-specific DNA binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGRKKISIAP ITDDRSRSVT FVKRKQGLYK KAYELAVLAD CEVAVTVIDR KGRLHVFCSS 60
61 DYQRTLQQLN TLSIYELKNR SHFSSSPVEE SSTVSPETTT GSFTPLNNKH LKSQDQPLSD 120
121 SQLDTGDSPA TSETTVQDYN PQVQSYCRPE PLSSNHVRSC PPFPPTQHHH PHTRPPHHPP 180
181 HPHFHNNNYP PPYCFQSPVS PGATVPLQHH SPYPSDNGFP GHRRQTHFAP YYYPQRATSP 240
241 SLKQVPTYLG THVLQQDQST YDQKVLMPPA SYLSSPNQYT LKNVSPGNPA CPPFLYEHPN 300
301 PQLTPEMFDV KQGSPIPPTA YSGSSCETSQ HTIANTPFLA YDRSPSLTNQ EAESSFQDVA 360
361 SISPHSLSDV KY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
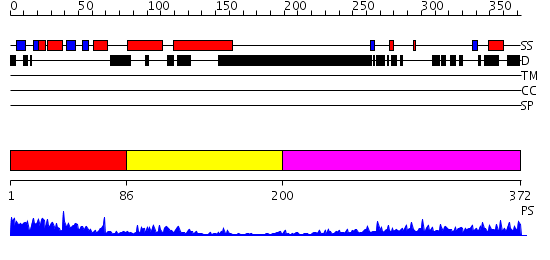
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | 29.221849 | Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2 | |
| 2 | View Details | [86..199] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [200..372] | 29.372997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)