













| General Information: |
|
| Name(s) found: |
pku80 /
SPBC543.03c
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 695 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromosome, telomeric region
[IDA nucleus [IDA cytosol [IDA |
| Biological Process: |
S phase of mitotic cell cycle
[IMP telomere maintenance [IMP double-strand break repair via nonhomologous end joining [IMP |
| Molecular Function: |
ATP binding
[IEA]
telomeric DNA binding [IDA protein heterodimerization activity [ISS ATP-dependent DNA helicase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDKECTVFV LDLGKDMGTC HHGRSHSDLE WTLSYFHDEL SHKFLANRKT DVVGIVGYKC 60
61 DDTKNDLAEQ EAYWNISVLY PIQTALFSKL QSVSQTLKPS NTMQGDLISA IVVSFDLMAR 120
121 HCKKNKWKKK MIVLTAARGI IDFSDYIGIA EQLLQHDVFL GVYGVDFDQE DINYSEPLKE 180
181 SQKKENEVRI QEFVESCHGQ YCTFQQIYNN IGKPWVRKVR PVAIFRGTFS IGNRDSKDTS 240
241 ISIQVERYPR TRLTKPPTSS AFYENDMSKN YECLNIENSN VENKSMESDA VSTVRSYMVR 300
301 DPKTNDSFEV KREDLESGYS YGRTIVPISR SDEDVLALDT IPGYEILGFI PKSSLPIYYT 360
361 ISDTNIIVPK DDFESKLNFS AFVQSLEREH RYALARFVSK DKGVPVLLVL MPYVEFKRHY 420
421 LVDIQLPFAE DVRPYSFSEF EKLSNEEDMR QIDFAVSNYI DNMDLDSSDC GFNPPFEPEN 480
481 TFSMIPHRLQ QAISYYANSP EGDLPQPNIY LTRYTNPPKS LLDNCILDLK LIKEKLTVNV 540
541 PVKPKYSSQE TAFDTGAPIS EEQIEELLNS GLDEQEGEKL LVLHVSEKDP VGTFTEVLKN 600
601 PFGLEDALTE MEKVIKNLID KSKYDLALQS LQSLRLHSIL EDEVERFNEY LTRLKKDVMQ 660
661 NNKPKENELI NKIRSSGLDI ILHDELTRHD NFNNI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
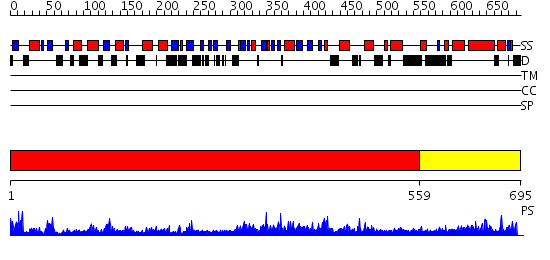
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..558] | 119.0 | Ku80 subunit | |
| 2 | View Details | [559..695] | 35.0 | Three-dimensional structure of Ku80 CTD |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)