













| General Information: |
|
| Name(s) found: |
SPAC17G6.05c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 775 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[ISS cytoplasm [ISS vacuole [RCA |
| Biological Process: |
protein targeting to vacuole during ubiquitin-dependent protein catabolic process via the MVB pathway
[ISS ubiquitin-dependent protein catabolic process [ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKLATPFFY LNKKETKHSD WVEPFTTFVS RIYGNSVDVE DQIKAFNTLR ENAADVDDTV 60
61 AGKDILYSYY GQLDYLSFRF PTGGNGINIS FEWSDILDPD ADFVKQSSLA FEKASVLFNL 120
121 VSLLSRMAAN HASAYTVDDY KAAANCLQCA SGIAKLLRES FIHAPGRDLD SNFLLGIYNL 180
181 FLGQAQECVL GHMSFSASDS NMNYSLAAKI ASSAATLYDS CVHAFESMEP ACNPNFIRLA 240
241 SAKKAALEGF SSYFMARAQL EKSKQGLAIG YLQQAKSILS SAQKLFNGIK LSTDFIHKPS 300
301 LSDYPQTIST FIKSSLSHLL KTAEKDNDFV FHDLVVKEVE LPKISPLQAL PPLPLEKLYG 360
361 SQDGFETAKK IVGDDLFKAF VPSAVTTASS LYSEETAKVF RAEQAEVERQ NTQMATVFAS 420
421 LDLSRLQEIT HSDSKTNFIP KELIEARSQL ISFNPTFKIH ELFAKSTKLK QIIAKIKSDL 480
481 EVEANENAKM KAKLGPSWTL SDSLEFAAPY QSELNNYLKT LAEASATDSA ISQQYALVKD 540
541 DVETLCNSSK ISALLESSIS STDNSGQSLL DIPIEQEEQE RDMKIQLDLL EELQSRVQKL 600
601 VPERQTTLQA LQQKCLQDDI SESLMQNSKR KDSALDTNQL FELELKKFDP LRNRLHASYR 660
661 QQQLLLNEMR NVTQKLKQNP EFLRKMEVYN NQFSKNKELC SNLLSSSLTA KAISEGVSKG 720
721 LEYYKTVEQR LAEINKTLGE QLLLRRKQGA TCLSKLSSSS SSHTSISSNM RNLHI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
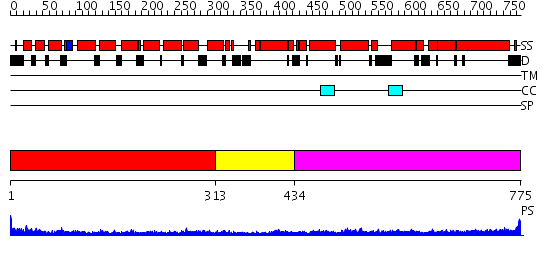
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..312] | 7.98 | Structure basis for endosomal targeting by the Bro1 domain | |
| 2 | View Details | [313..433] | 6.69897 | Tropomyosin | |
| 3 | View Details | [434..775] | 11.154902 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)