













| General Information: |
|
| Name(s) found: |
sol1 /
SPBC30B4.04c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 865 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SWI/SNF complex
[IDA nucleus [IDA nuclear chromatin [IC spindle pole body [IDA |
| Biological Process: |
chromatin remodeling
[ISS regulation of transcription from RNA polymerase II promoter [IDA |
| Molecular Function: |
DNA binding
[ISS general RNA polymerase II transcription factor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNQGFVPAS DYPTAVSYPT QGQSYNTQEE QPAYPQRFST SQGMYAAEYG NANMMNTSEN 60
61 EPNNLAHSQP FRQSPSTQRN LPNQSFDFAS NGAWNGSGSV KYSSPMMPSS RIPFQQEKEA 120
121 AMQQQQQQQQ QQQLYQRQMQ SREALLSQQI PPNQIGINAH PAVRQTPQPA PSPNTPSGNA 180
181 NQLTPAYAAS FDKFMVSLIS FMEKRGTPIK SYPQINNTPI NLMMLYALVM RAGGSRQVSA 240
241 HNFWPKISAS LGFPSPDAIS LLIQYYNSYL LPYEEAWLAA QQQQKSLQQA KANHSANVQS 300
301 RPKNYPQKPV QTTPEAVHAN GSMHGSLHSK SPSPAFTANR FSPAAPTTVS SERNAPPYPS 360
361 APTRPTPPTV QTSSSAAPVD SAEPVAYQPI KKPIDPMLGY PLNVAATYRL DESLLRLQMP 420
421 SIVDLGTVNI QALCMSLQST LEKEITYAMN VLLILTNDQK WMFPLSECQD VVDALIDVAT 480
481 QCLDNLLSVL PNEDLMEIAD KRPSYRQLLY NCCVEISQFS REDFSNSLSE NKTKDSINAI 540
541 DVHNSEQNLL AVFVIFRNLS HFEANQNVLV QNPDFFPLLI RVVKSLNFHA TSLLRSSRNT 600
601 LDLHKDVLIV LCQLSQNFIL PNVDVARHVL LFILSFSPFN RKKSKTILND TLPTSIPSYT 660
661 PATHPYAGPA INAYAKLLAK DANNKTNFQA IFDNNPKFLD SLFLLLASVV PKFNRHCLKI 720
721 CERRLPLLQQ SFFCLAATVS YVKQSEQAAN WCNIGEGFFV SMLRLLILLS GHPSLNPPSR 780
781 VASQYPTTNP FRYVIQSGIS TVRRLLSLVE AGNISLSSFP KSETLLAVLL APTTETSFLK 840
841 EISNLLDRTG DSDASLENTD DKSGI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
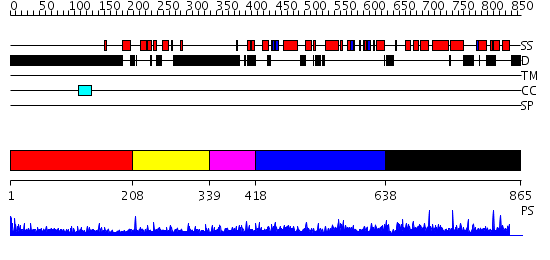
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..207] | 6.154902 | Sec24 | |
| 2 | View Details | [208..338] | 19.0 | DNA-binding domain from the dead ringer protein | |
| 3 | View Details | [339..417] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [418..637] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [638..865] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)