













| General Information: |
|
| Name(s) found: |
SPAC6G9.14
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 681 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[ISS cytosol [IDA |
| Biological Process: |
nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay
[ISS |
| Molecular Function: |
mRNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEQSSKSHP SFMSTEYSLP VFSSQYSDIE NKDVLKSNSW DPYAANAPIA LSEIPSSSKS 60
61 GKTNGSKAAV ESGLASGAFG NRTGNMNRNT SMSGYTFGSS NFVPYNLLSN TPSFTTSHSS 120
121 STTFVPPATM GGGLNNLSSP SSSLYLPGSA SYQRSNSKNS SASILQMNTT LDDIPILLRR 180
181 PGLSSYTPGP STSRRSISSS SNLGGNPGLI ANNPSASKNF AFTSGSSSTN NSTTSSSSMA 240
241 NGLQSISKHS AAFPLLSNAT SFFGENLTPS LAASTASTGS TDSSGSNIAN TLIAPTPTIS 300
301 PPSANTVGNP SPADTPGFNV PSLISDDPSV SSSLSSSVAS LSLQNSNILS FCKDQHGCRY 360
361 LQRLLEKKNQ SHIDAVFAET HPYLAVLMVD AFGNYLCQKL FEHASEAQRS TFIQIIAPKL 420
421 VPISFNMHGT RALQKIIDLV SSPDQISCIV NALRPNVVLL TKDLNGNHVI QKCLNKFSQE 480
481 DCQFIFDAIC EDPLDVSTHR HGCCVVQRCF DHASPAQIEQ LVEHIVPHAL TLVQDAFGNY 540
541 VLQYVLELNN PNHTEAIISY FLYKVRALST QKFSSNVMEK CIFFAPAAIK EKLISELMDE 600
601 KHLPKLLRDS FANYVIQTAL DNASVKQRAE LVERIKPLIP SIKNTPCGRR ILSKLERRHP 660
661 SSKEKPIVYS NSERVNTSSS A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
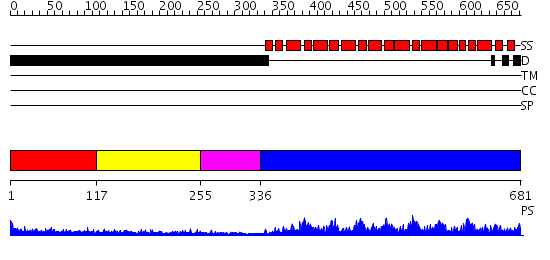
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | 3.416996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [117..254] | 3.467997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [255..335] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [336..681] | 135.0 | Pumilio 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)