













| General Information: |
|
| Name(s) found: |
ppk2 /
SPAC12B10.14c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 665 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell tip
[IDA cytosol [IDA |
| Biological Process: |
signal transduction
[RCA protein amino acid phosphorylation [ISS |
| Molecular Function: |
ATP binding
[ISS protein serine/threonine kinase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSNSTFHEH HAKSHFHNNA CQSNASSSAC RASEDHLVSS FPNDSIIDLQ PSRPAPEPPK 60
61 KKFGYYARRL SGHFLSLIHG SGNSTRSPPF HLQNQKSNGQ SEVWHSSDDS GSPKRLNRSR 120
121 SSKEDMYRRR SLHGLPSLSR RNSKKSSTLS RSISLHLRSE SAPISLPIHL YKSYSYNHSP 180
181 SSLPTVLNSQ ALSSPPVPTT PDEVSTNRLS SSTSSMNCRN LVPDNFNISI RPNTTNYRSS 240
241 IQENSNGNRD SISPSAYDAP LLHNVDTQSI DGFVSVASHF SSASTAESLD DGHSATTIQQ 300
301 GDVSSYPLSR SVSTPVPMSP ISISPAKPSP QSPKLSQSAV GHPSSSIPAA AMHKVSYSDD 360
361 LMRFVAREKY YLQIVDCLCT QKDPLFFYTD FTKICQQDTV GTYVARQTLD KEVVVIKRFD 420
421 ISAVTHRRLL LEELQRLSGL SHKNLIRYNE SFWYLNNIWS VFEYKDPSTK LSALIPKYFF 480
481 SELNIASICY EISSGLAFLH NSGIAHHNLT TECIYLTKSS CLKIGNYAFS SPYIERQTNR 540
541 GAVSHVPDWL IEKNYKEGFM KDVKSLGLVA LEIFQGQPNF FRKSIQSIQL TPNANVLVNR 600
601 VRGLISQEFK EFLLQTLQAE TLQGPNINML LETSSFLEKR QTLNFEICLN NLNLRERKAS 660
661 RYSYL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
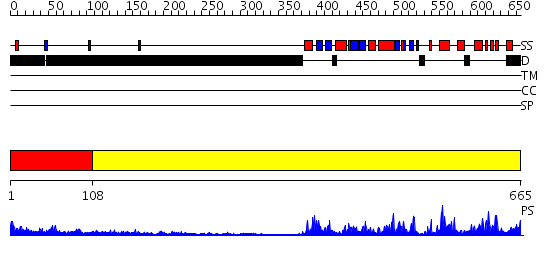
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [108..665] | 75.30103 | No description for 2h8hA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)