













| General Information: |
|
| Name(s) found: |
set3 /
SPAC22E12.11c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 859 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Rpd3L-Expanded complex [IDA Set3 complex [IDA |
| Biological Process: |
chromatin remodeling
[IPI histone methylation [IC |
| Molecular Function: |
zinc ion binding
[IEA]
histone-lysine N-methyltransferase activity [TAS small conjugating protein ligase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWKIRCVCPF EDDDGFTIQC ESCEVWQHAV CVNIDANNVP EKYFCEQCQP RPIDADKAHK 60
61 IQLARLQREE EQSRILSRSR SSNNKRRTSF GKNGASPTHS ASPRQGNNTG ANGALFSQST 120
121 NSSNSGSYRN SVTGATLPNA HAPHSQNRRR RSNHLNNPPE APITEASNEY VYSFHLEYVP 180
181 LESNTFSASA LEYSKNLDLK NLDESEVLMD GCQVVPISSS KFCCSRFGLV STCEIPPNTP 240
241 IMEVKGRVCT QNEYKSDPKN QYNILGAPKP HVFFDSNSQL VVDSRVAGSK ARFARKGCQS 300
301 NSVVSSVYMN GSNSVPRFIL YSTTHIAPET EIIGDWTLDI SHPFRQFAPG MSRPSFNMEE 360
361 LELLSEVLST FLSFNECASQ DKKNCVFSRV TKYIKAARRA STANRVSVAK DRLSLTPSST 420
421 PSTPSPAESL PQPSNPTSVY AKSLKEFWLD KYRLSILQKW PAVKSLPTES VGIDVVMEPK 480
481 LQPSVKEKKP TKDLQSPLPS VEEDSSNRDK KTDIADLHTD SKVGIADVLS PISPDAALQS 540
541 DGPLKKAKEP EESSITPTTP PSFNVGESLS RRSASPLQHP RTSPDMLDKT SPCKRGLGTI 600
601 TTVHKKHGSV DHLPSVKRRR SIANDFHGKP DYNKRSLSIE RKPEAFKTKG DRPHKVHPSF 660
661 HRNSDSKLKL EPSSKEKSGS MFFNTLRTVK DKSHVHDTQR SSDVNFSRQN GTRSHSPSVS 720
721 PVGFSFDKSP VTTPPLPTAP APVITSRHAL VNNQFPTNNP NILDHKANNG DDISNALNTS 780
781 RSENKPNSNL VQGSVVKPSN TSASALPTSA PKKLSLSEYR QRRQQNILHQ QSKDNQAHGD 840
841 TARPHTVPAA TVSNPSFTR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
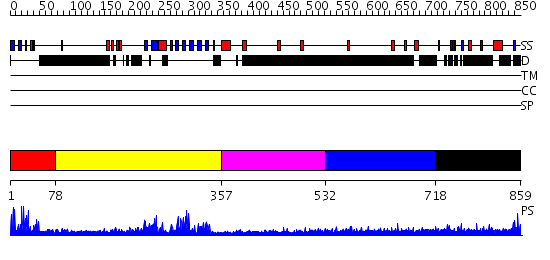
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | 14.522879 | Solution structure of PHD domain in death inducer-obliterator 1(DIO-1) | |
| 2 | View Details | [78..356] | 57.045757 | No description for 2igqA was found. | |
| 3 | View Details | [357..531] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [532..717] | 1.232997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [718..859] | 3.228994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)