













| General Information: |
|
| Name(s) found: |
phx1 /
SPAC32A11.03c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 942 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [NAS |
| Biological Process: |
regulation of transcription, DNA-dependent
[IGI gene-specific transcription from RNA polymerase II promoter [RCA |
| Molecular Function: |
DNA binding
[TAS transcription factor activity [TAS specific RNA polymerase II transcription factor activity [RCA sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRSYSNPENG GQINDNINYS EKRPTMLPEN LSLSNYDMDS FLGQFPSDNN MQLPHSTYEQ 60
61 HLQGEQQNPT NPNYFPPEFD ENKVDWKQEK PKPDAPSFAD NNSFDNVNSS KLTNPSPVQP 120
121 NIVKSESEPA NSKQNEVVEA TSVEKAKENV AHESGTPESG GSTSAPKSKK QRLTADQLAY 180
181 LLREFSKDTN PPPAIREKIG RELNIPERSV TIWFQNRRAK SKLISRRQEE ERQRILREQR 240
241 ELDSLNQKVS QAFAHEVLST SPTSPYVGGI AANRQYANTL LPKPTRKTGN FYMKSGPMQS 300
301 SMEPCIAESD IPIRQSLSST YYNSLSPNAV PVSSQRKYSA SSYSAIPNAM SVSNQAFDVE 360
361 SPPSSYATPL TGIRMPQPES DLYSYPREVS PSSGGYRMFG HSKPSSYKAS GPVRPPNMAT 420
421 GHMRTSSEPT SYDSEFYYFS CTLLVIGLWK RLRASPQDLM CFYSPPKKLF AYLIQFQGIQ 480
481 YRIEYSFFVI ESIHVFRVEE PLLNELSATA SSRDKPAPNE YWLQMDIQLS VPPVFHMITS 540
541 EGQGNCTDFT EGNQASEVLL HSLMGRATSM FQMLDRVRRA SPELGSVIRL QKGLNPHQFL 600
601 DPQWANQLPR QPDSSVFDHQ GRNPPIQGLS HDTSSEYGNK SQFKRLRSTS TPARQDLAQH 660
661 LLPPKTNTEG LMHAQSVSPI TQAMKSANVL EGSSTRLNSY EPSVSSAYPH HNLALNLDNT 720
721 QFGELGTSNI SYPLSAPSDV GSLPRASNSP SRPVMHPNTQ GINTEIKDMA AQFPNSQTGG 780
781 LTPNSWSMNT NVSVPFTTQN REFGGIGSSS ISTTMNAPSQ QLSQVPFGDV SLATENSVPS 840
841 YGFEVPSEES VYAQARTNSS VSAGVAPRLF IQTPSIPLAS SAGQDSNLIE KSSSGGVYAS 900
901 QPGASGYLSH DQSGSPFEDV YSPSAGIDFQ KLRGQQFSPD MQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
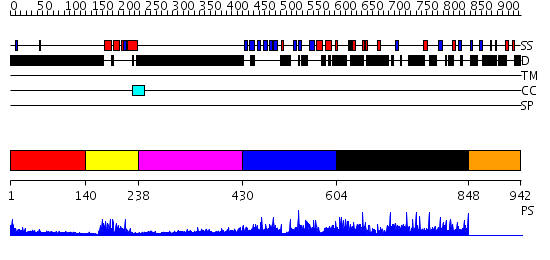
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..139] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [140..237] | 18.0 | Homeobox protein hox-b1 | |
| 3 | View Details | [238..429] | 4.665997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [430..603] | 1.142968 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [604..847] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [848..942] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)