













| General Information: |
|
| Name(s) found: |
ubp12 /
SPCC1494.05c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 979 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
protein deubiquitination
[TAS ubiquitin-dependent protein catabolic process [TAS |
| Molecular Function: |
ubiquitin thiolesterase activity
[ISS cysteine-type peptidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSLSESSTS SYHGKRPRSL SEESQSSSNM DDISQKSISL GDASEISKNL PSIAEQKQLI 60
61 GELVNNQPEL ELGQVDNYIL SYSWYERLCS YLAEDGPFPG PVDQEDIADL ETGTLKPDLQ 120
121 EEIDFTIISR DVWDLLVRWY GLKGPEFPRE TVNLGSESHP HLVVEVYPPI FSLTLLSTNA 180
181 VDANESHKPK KISLSSKSTL EDLLEGVKYT LSLPSDQFRL WRVDTDQPLH RTIDPSSFIK 240
241 INSKEIIDFL EKSKTLVELG MDSSCSLVAE CMINETWPVD RALRLQFLIQ QRNNQSSNEE 300
301 QKQEKRVPGT CGLSNLGNTC YMNSALQCLT HTRELRDFFT SDEWKNQVNE SNPLGMGGQV 360
361 ASIFASLIKS LYSPEHSSFA PRQFKATIGK FNHSFLGYGQ QDSQEFLAFL LDGLHEDLNR 420
421 IYQKPYTSKP DLYEVDEEKI KNTAEECWRL HKLRNDSLIV DLFQGMYRST LVCPVCNTVS 480
481 ITFDPFMDLT LPLPVKQVWS HTVTFIPADT NLTPLAIEVV LESKAATIED LVKYVAEKSG 540
541 CSDYRKILVT ETYKGRFYRF LTQLSKSLLM EISEEDEIYL YELERPYEDG SDDILVPVYH 600
601 ISDDSTNSAN SYMSSRDFGH PFVLQLSDNE VTDASFISEK LKLKYQQFTT LKNLKNIDSL 660
661 ESLELGHEDE QVQKGPLDVD MDHSQTPLFE MRVFHDRFEK IPTGWNMSVS NLPLLTERDK 720
721 KDLESTVDPL DAHSIEEEDD SEFKDVAPGS YPEPSKSNEN TKLTAKENDR LLIQGDLLVC 780
781 EWPEKSYQFV FSVAPSSPQM GRSLWLESKT ILSDKKDDSE DSRTITLNDC LDEFEKTEQL 840
841 GEEDPWYCPT CKEFRQASKQ MEIWRCPEIL IFHLKRFSSE RRFRDKIDDL VEFPIDNLDM 900
901 SMRTGSYKLS EKENPKLIYE LYAVDNHYGG LGGGHYTAFA KNPDNGQFYC FDDSRVTPVC 960
961 PEETVTSAAY LLFYRRKTS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
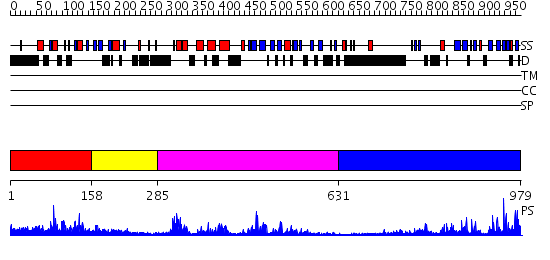
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | 28.39794 | Solution structure of the DUSP domain of hUSP15 | |
| 2 | View Details | [158..284] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [285..630] | 61.221849 | No description for 2gfoA was found. | |
| 4 | View Details | [631..979] | 36.09691 | Ubiquitin carboxyl-terminal hydrolase 7 (USP7, HAUSP) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)