













| General Information: |
|
| Name(s) found: |
ste20 /
SPBC12C2.02c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1309 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA TORC2 complex [IDA cell division site [IDA cytoplasm [IDA integral to membrane [IEA] cytosol [IDA |
| Biological Process: |
small GTPase mediated signal transduction
[ISS TOR signaling pathway [TAS cellular response to nitrogen starvation [IEP regulation of conjugation with cellular fusion [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKPVRRGQTD TALDISSHAK TNGDFIKKMN TTDSKRLKLL EDLKGKLEVE CKIRDGAETL 60
61 LQVFDTNFKK ETKERKEMLK KKCTDELESS KKKIEELVSS IESFQGENGE AKTGSTSLTR 120
121 SASATVSRKS SLQEKYSTRF SYKAGCSDSC SVTVSGTGEL IGPTRNAHSN LTPTVIQRID 180
181 FENVNEKNNS SSEDTQPNGK RPSSLQSNFS QFPLNPWLDN IYKACLEGSM KDVIDSSNNL 240
241 CEYLHEHSDP AYAKNFSLIT PTILSMLELN VSEVTASVYR LLRHLFLDAT AFSCCQMLNL 300
301 PWILSKSLLS GTDAYQIERE QAFRLIRTLY FLSSTEGHED YLSGITRTII SICEHVSDVS 360
361 RGIAVETLIE LMIIRPKILF KANGLRVLMI SLIDGSISEN LAASAALALV YLLDDPESAC 420
421 YVNLPYDIGI LLSPFTSSSS RDTFNSSEEQ SEQAAKAMKS SAKVASVLLN SWSGLLALST 480
481 NDFQALRSIV DTLRVPSFAP RSDVIDLFFL IFQVEYSSWS ESFLAGKRLT VVKNQAVSND 540
541 DNINMVNIPD GSNKKYMSLR QHFTAVLLFI FLELGLVESI VCMIRASDDP SASRKATYLL 600
601 GEVLRLSDEL LPIHLGAKIQ SLPSLFNMAS QFTAEDRFVA TSVLQSIESL NRVKFHSATQ 660
661 PFSQTTSLLF KEQKTDGSFR GQRQVEHVKL KMGMQIDDSH FRSMLAETNV LATKNYQKWR 720
721 WDTLVQIMEG PLLSPKRIDE TLRTTKFMRR LLAFYKPFSN RFSSIQNTKP NQKFIKVGCL 780
781 VFRTLLANPE GVKYLSESKV IKQIAESLSQ IDGYSEQVSE PIFSNSRLQK TLTHGYFPML 840
841 KVLSSQKEGH AIMERWRIFT TLYHLTELRN RDDLIIIFLT NLDYRLEGHT RIIFSKALNT 900
901 GQQAVRLTAT KHLAALINSE SANDNLNHWA ISLLIFQLYD PCLEVCKTAV KVLNEVCARN 960
961 ENLLAQVVQL QPSLAHLGEI GSPLLLRFLA TTVGFHYLSE INFIEHELDN WYHHRNIDYV1020
1021 DLLEQNFFLS FVSNLKIIDK KNNEPDENIL PLHFYGELVK SPQGCEVLES SGHFESFMGT1080
1081 LVEFYDKPLG NEAIRQLKSA LWAIGNIGKT DQGITFLINH DTIPLIVKYA ENSLIPTVRG1140
1141 TAYFVLGLIS RTSKGVEILE SLHWYSLMSL MGTSQGICIP RHAGQVLSTP RRNVEFVNER1200
1201 VPTPEFSSLL SSLTNSEREV IRLVSNLSNH VLTNESARQL TKIRSKNAKV FSSKRLVKAC1260
1261 MTILGKFHYR VQIQQFVFEL FPYSVLLSSS TSQDLNESPS RPNNLSISA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
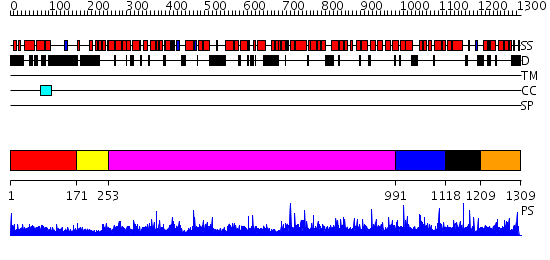
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..170] | 13.69897 | Structure of a helically extended SH3 domain of the T cell adapter protein ADAP | |
| 2 | View Details | [171..252] | 1.02 | Crystal structure of the HspBP1 core domain | |
| 3 | View Details | [253..990] | 1.08 | No description for 2ie3A was found. | |
| 4 | View Details | [991..1117] | 1.04498 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1118..1208] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1209..1309] | 1.020978 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)