













| General Information: |
|
| Name(s) found: |
rec8 /
SPBC29A10.14
[Sanger Pombe]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 561 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA inner kinetochore of condensed chromosome [IDA nuclear chromosome [IC cohesin complex [IDA meiotic cohesin complex [IDA |
| Biological Process: |
attachment of spindle microtubules to kinetochore involved in homologous chromosome segregation
[IMP linear element assembly [IMP reciprocal meiotic recombination [IMP meiotic sister chromatid segregation [IDA homologous chromosome segregation [IDA meiotic DNA double-strand break formation [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFYNQDVLTK EKGGMGVIWL AATLGSKHSL RKLHKKDIMS VDIDEACDFV AFSPEPLALR 60
61 LSSNLMIGVT RVWAHQYSFF HSQVSTLHLR VRKELDHFTS KPFKNIDIQN EQTNPKQLLL 120
121 AEDPAFIPEV SLYDAFNLPS VDLHVDMSSF TQPKENPNIS VLETLPDSTS YLINTSQNYS 180
181 LRNNVSSFVY EDSRAFSTEE PLDFEFDENG DIQELTKGTI NSDPSLQAAS QHSNLGSVQR 240
241 EYNSEEQESR IHMFEIDEDV LPLPVPLQSV MDSEHNENEP RALKRRKVQK LLEPDENIEL 300
301 STRTLSQWRK NYVERMIALE ATKYVRRRGA SSAKKKELNK FFDWESFHPL LKPWIEKLKP 360
361 SNNTPSEIDD VLRNIDTSEV EVGRDVQGEL GLNIPWNTSS RSNSAINSKS HSQTGSEHST 420
421 PLLDTKYRKR LPHSPSMPSR VEFLPALESS QFHETLNSEL SLQLSDDFVL YKNTQEENAH 480
481 LMLSMEKECA NFYEYAKTAI YENNGRITFS SLLPNDLKRP VVAQAFSHLL SLATKSAFLV 540
541 KQDKPYSEIS VSLNLKSTDA I |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
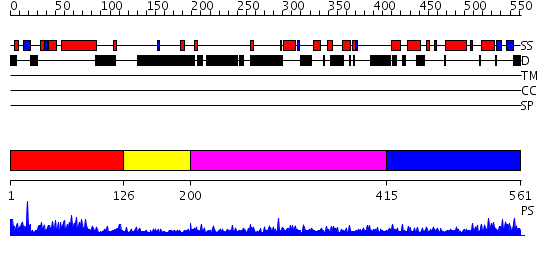
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..125] | 57.958607 | No description for PF04825.4 was found. No confident structure predictions are available. | |
| 2 | View Details | [126..199] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [200..414] | 1.090988 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [415..561] | 11.69897 | Sc Smc1hd:Scc1-C complex, ATPgS |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)