













| General Information: |
|
| Name(s) found: |
reb1 /
SPBC1198.11c
[Sanger Pombe]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 504 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS nucleolus [RCA |
| Biological Process: |
replication fork arrest at rDNA repeats
[IMP termination of RNA polymerase I transcription [IMP regulation of transcription termination [IC |
| Molecular Function: |
rDNA spacer replication fork barrier binding
[TAS RNA polymerase I transcription termination factor activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTSVLNPEL QIHGFIGVDS LQSSRKRKND FDDFPLNKGL KTNNNDYSGS IEPKFSPALS 60
61 IKEDGKNDRN FEALMSLQAQ DSNLSEQNTS IHLDALASSS IALGNDNVDS SAFLSKVNKG 120
121 VNAMRNSTSN QTNDSILISP SEITNMDPFL KGSARWTAEH WDYLERRMQN FCQTYSLDHT 180
181 QVADSLHEKR LHGPLSSLVK LLVQEMPSFT RRTILRHLRA LYNIPGYEKY SRKNSSGRGD 240
241 FGVQETAIIS QEVHNFIMDQ GWSEYQFCNQ IWAGKCPKTI RMFYSNLYKK LSHRDAKSIY 300
301 HHVRRAYNPF EDRCVWSKEE DEELRKNVVE HGKCWTKIGR KMARMPNDCR DRWRDVVRFG 360
361 DKLKRNAWSL EEETQLLQIV AELRNREDLS SDINWTLVAQ MLGTRTRLQC RYKFQQLTKA 420
421 ASKFELQENV WLLERIYDSL LNNGGKIHWE NIVKEANGRW TRDQMLFQFI NLKKMIPSYD 480
481 NLPLLEATKS AIDDFKVVLS GFSN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
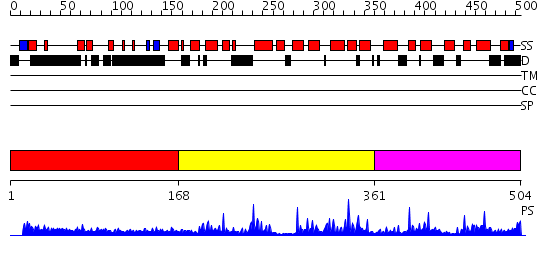
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 2.079983 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [168..360] | 36.154902 | c-Myb, DNA-binding domain | |
| 3 | View Details | [361..504] | 4.26 | c-Myb, DNA-binding domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)