













| General Information: |
|
| Name(s) found: |
pzh1 /
SPAC57A7.08
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 515 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA endoplasmic reticulum [IDA |
| Biological Process: |
regulation of translation
[ISS]
cellular potassium ion homeostasis [IMP cellular water homeostasis [TAS signal transduction [RCA regulation of ATPase activity [IMP protein amino acid dephosphorylation [IC cellular sodium ion homeostasis [ISS response to cation stress [TAS |
| Molecular Function: |
protein serine/threonine phosphatase activity
[TAS iron ion binding [IEA] manganese ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGQGSSKHAD SKLDSYPSFS RSDTQGSIKS LKSLKTVLGK GKDSNHDRRT STDTTHSRHR 60
61 YPETPPSLPP PPSPGILATS PAVLQKHQQE DSGNSSQSPT SPHPSNQPAM LSPSTAASQH 120
121 HHHHSSSSSY AVSPTSPTSP TSSGPIGSNF DSASEHNGPV YPQDQQGPVI IPNSAISSTD 180
181 PDDPETVVSL NVDEMIQRLI HVGYSRKSSK SVCLKNAEIT SICMAVREIF LSQPTLLELT 240
241 PPVKIVGDVH GQYSDLIRLF EMCGFPPSSN YLFLGDYVDR GKQSLETILL LFLYKIRYPE 300
301 NFFLLRGNHE CANITRVYGF YDECKRRCNI KIWKTFINTF NCLPIASVVA GKIFCVHGGL 360
361 SPSLSHMDDI REIPRPTDVP DYGLLNDLLW SDPADTENDW EDNERGVSFV FNKNVIRQFL 420
421 AKHDFDLICR AHMVVEDGYE FFNDRTLCTV FSAPNYCGEF DNWGAVMSVN SELLCSFELI 480
481 KPLDQAAIRR ELKKSKRSGM AIYQSPPAEQ VTQSV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
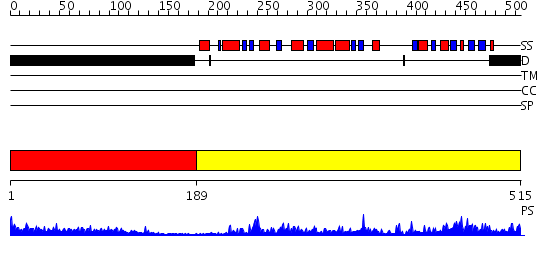
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..188] | 1.24 | No description for 2j63A was found. | |
| 2 | View Details | [189..515] | 103.0 | Complex between protein ser/thr phosphatase-1 (delta) and the myosin phosphatase targeting subunit 1 (MYPT1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)