













| General Information: |
|
| Name(s) found: |
pms1 /
SPAC19G12.02c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 794 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromosome [ISS |
| Biological Process: |
maintenance of DNA repeat elements
[IMP mismatch repair [IGI |
| Molecular Function: |
ATPase activity
[ISS ATP binding [ISS DNA binding [ISS mismatched DNA binding [NAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTVKPIDAN TVHKICSGQV ITDVASAVKE LVENSLDSGA TTIEIRFKNY GINSIEVVDN 60
61 GSGIDAGDYE SIGKKHFTSK ITDFEDLEAL QTFGFRGEAL SSLCAVGQVI ISTATQNEAP 120
121 KGVQLNLDHE GSLKDKLTIP FQRGTSVMVN DLFCTLPVRR KLLEKNYKRE FSKAISLLQA 180
181 YATISTNKRF MVYHQTKNSG KLLQLSTNSN KDMKLNIMNV FGTKVSSSLI PWNDGIIEGY 240
241 ISRPHVGSTR ASNERQMLFI NRRLVNLPKI ARVIQEVFKP YSMAQSPFFA INLRITNGTI 300
301 DINVSPDKKS VFLSEEDSII EFIKNSLQNL CESCGHAISC SRSQSIFSYS SQIPDSSGDS 360
361 TDQELPQSIP ATESETSDDS SFSYKRSPCK RKLVEATAQP AISTSVAEGA SLAQVSKPLP 420
421 ERLQKDSMRR SSPLNEKVTA SSERMKKKLA LFASSTDTSM QKTIDSSFPL KQPINKPSSN 480
481 PNNLLLNDPS PASTPVAKTI NLNEIESVHN AESVSTLSSI PRTEQTSVAN RIPSKTAALQ 540
541 KLKFFQSRPL DGLNKFSKKI NISLSGVQKD IVRSDALLKF SNKIGVVHDI SDENQEDHLN 600
601 LTVHKADFLR MRVVGQFNRG FIVVVHGNNL FIIDQHASDE KFNYEHLKSN LVINSQDLVL 660
661 PKRLDLAATE ETVLIDHIDL IRRKGFGVAI DLNQRVGNRC TLLSVPTSKN VIFDTSDLLE 720
721 IISVLSEHPQ IDPFSSRLER MLASKACRSS VMIGRALTIS EMNTIVRHLA ELSKPWNCPH 780
781 GRPTMRHLLR LKDI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
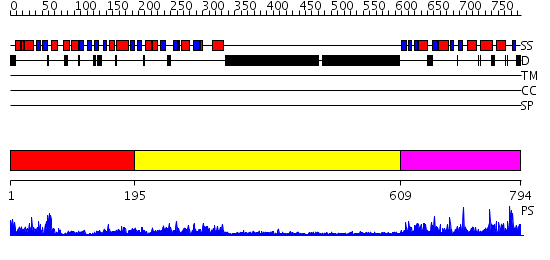
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..194] | 61.522879 | DNA mismatch repair protein PMS2 | |
| 2 | View Details | [195..608] | 26.221849 | No description for 2cgeA was found. | |
| 3 | View Details | [609..794] | 22.30103 | Crystal structure of the MutL C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)