













| General Information: |
|
| Name(s) found: |
pka1 /
SPBC106.10
[Sanger Pombe]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 512 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cytosol [IDA cAMP-dependent protein kinase complex [TAS |
| Biological Process: |
induction of conjugation upon nitrogen starvation
[IMP negative regulation of transcription by glucose [IMP age-dependent response to reactive oxygen species [IGI protein amino acid phosphorylation [IC negative regulation of conjugation with cellular fusion [IMP cAMP-mediated signaling [IC chronological cell aging [IGI cellular response to nitrogen starvation [IMP |
| Molecular Function: |
ATP binding
[ISS protein binding [IPI cAMP-dependent protein kinase activity [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTTAVASKG STNVGSSTDT LSTSASLHPS MNAGSVNEYS EQQRHGTNSF NGKPSVHDSV 60
61 GSDASVSNGH NNHNESSLWT SGIPKALEEA TKSKKPDSLV STSTSGCASA HSVGYQNIDN 120
121 LIPSPLPESA SRSSSQSSHQ RHSRDGRGEL GSEHGERRSA MDGLRDRHIR KVRVSQLLDL 180
181 QRRRIRPADH TTKDRYGIQD FNFLQTLGTG SFGRVHLVQS NHNRLYYAIK VLEKKKIVDM 240
241 KQIEHTCDER YILSRVQHPF ITILWGTFQD AKNLFMVMDF AEGGELFSLL RKCHRFPEKV 300
301 AKFYAAEVIL ALDYLHHNQI VYRDLKPENL LLDRFGHLKI VDFGFAKRVS TSNCCTLCGT 360
361 PDYLAPEIIS LKPYNKAADW WSLGILIFEM LAGYPPFYSE NPMKLYENIL EGKVNYPSYF 420
421 SPASIDLLSH LLQRDITCRY GNLKDGSMDI IMHPWFRDIS WDKILTRKIE VPYVPPIQAG 480
481 MGDSSQFDAY ADVATDYGTS EDPEFTSIFK DF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
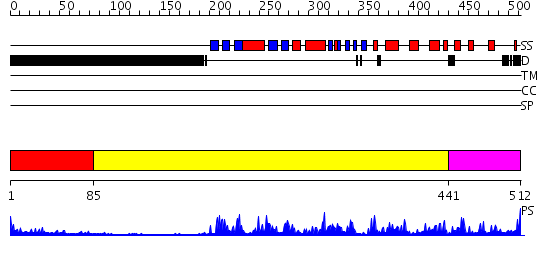
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 3.221849 | Polygalacturonase inhibiting protein PGIP | |
| 2 | View Details | [85..440] | 53.30103 | Hemapoetic cell kinase Hck; Hemopoetic cell kinase Hck; Haemopoetic cell kinase Hck | |
| 3 | View Details | [441..512] | 95.69897 | cAMP-dependent PK, catalytic subunit |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)