













| General Information: |
|
| Name(s) found: |
ssc1 /
SPAC664.11
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 674 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA presequence translocase-associated import motor [IDA |
| Biological Process: |
protein import into mitochondrial matrix
[ISS regulation of catalytic activity [IC cellular response to stress [ISS protein folding [ISS |
| Molecular Function: |
ATPase activity
[ISS ATP binding [ISS unfolded protein binding [IEA] enzyme regulator activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISSRFTNVV RSGLRFQSKG ASFKIGASLH GSRMTARWNS NASGNEKVKG PVIGIDLGTT 60
61 TSCLAIMEGQ TPKVIANAEG TRTTPSVVAF TKDGERLVGV SAKRQAVINP ENTFFATKRL 120
121 IGRRFKEPEV QRDIKEVPYK IVEHSNGDAW LEARGKTYSP SQIGGFILSK MRETASTYLG 180
181 KDVKNAVVTV PAYFNDSQRQ ATKAAGAIAG LNVLRVVNEP TAAALAYGLD KKNDAIVAVF 240
241 DLGGGTFDIS ILELNNGVFE VRSTNGDTHL GGEDFDVALV RHIVETFKKN EGLDLSKDRL 300
301 AVQRIREAAE KAKCELSSLS KTDISLPFIT ADATGPKHIN MEISRAQFEK LVDPLVRRTI 360
361 DPCKRALKDA NLQTSEINEV ILVGGMTRMP RVVETVKSIF KREPAKSVNP DEAVAIGAAI 420
421 QGGVLSGHVK DLVLLDVTPL SLGIETLGGV FTRLINRNTT IPTRKSQVFS TAADGQTAVE 480
481 IRVFQGEREL VRDNKLIGNF QLTGIAPAPK GQPQIEVSFD VDADGIINVS ARDKATNKDS 540
541 SITVAGSSGL TDSEIEAMVA DAEKYRASDM ARKEAIENGN RAESVCTDIE SNLDIHKDKL 600
601 DQQAVEDLRS KITDLRETVA KVNAGDEGIT SEDMKKKIDE IQQLSLKVFE SVYKNQNQGN 660
661 ESSGDNSAPE GDKK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
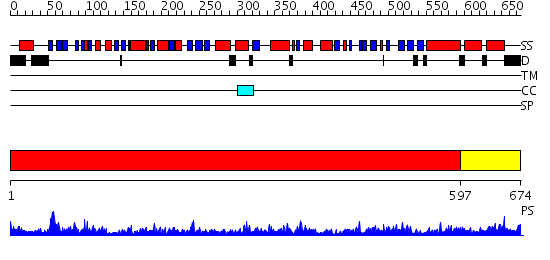
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..596] | 1000.0 | crystal structure of bovine hsc70(aa1-554)E213A/D214A mutant | |
| 2 | View Details | [597..674] | 59.221849 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)