













| General Information: |
|
| Name(s) found: |
esc1 /
SPAC56F8.16
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 413 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
induction of conjugation upon nitrogen starvation
[IMP positive regulation of transcription, DNA-dependent [IMP gene-specific transcription from RNA polymerase II promoter [RCA |
| Molecular Function: |
DNA binding
[ISS transcription factor activity [TAS specific RNA polymerase II transcription factor activity [RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSYALPSMQ PTPTSSIPLR QMSQPTTSAP SNSASSTPYS PQQVPLTHNS YPLSTPSSFQ 60
61 HGQTRLPPIN CLAEPFNRPQ PWHSNSAAPA SSSPTSATLS TAAHPVHTNA AQVAGSSSSY 120
121 VYSVPPTNST TSQASAKHSA VPHRSSQFQS TTLTPSTTDS SSTDVSSSDS VSTSASSSNA 180
181 SNTVSVTSPA SSSATPLPNQ PSQQQFLVSK NDAFTTFVHS VHNTPMQQSM YVPQQQTSHS 240
241 SGASYQNESA NPPVQSPMQY SYSQGQPFSY PQHKNQSFSA SPIDPSMSYV YRAPESFSSI 300
301 NANVPYGRNE YLRRVTSLVP NQPEYTGPYT RNPELRTSHK LAERKRRKEI KELFDDLKDA 360
361 LPLDKSTKSS KWGLLTRAIQ YIEQLKSEQV ALEAYVKSLE ENMQSNKEVT KGT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
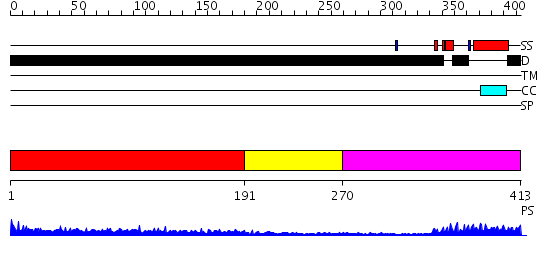
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | 5.133997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [191..269] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [270..413] | 22.0 | Myc prot-oncogene protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)