













| General Information: |
|
| Name(s) found: |
pap1 /
SPAC1783.07c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 552 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA NuA4 histone acetyltransferase complex [IDA cytoplasm [IDA cytosol [IDA |
| Biological Process: |
[NOT]response to arsenic
[IGI response to methylglyoxal [IMP cellular response to oxidative stress [IMP regulation of transcription, DNA-dependent [TAS response to caffeine [IMP histone acetylation [IC] response to hydrogen peroxide [IGI gene-specific transcription from RNA polymerase II promoter [RCA cellular response to nitrogen starvation [IEP] |
| Molecular Function: |
transcription factor activity
[TAS specific RNA polymerase II transcription factor activity [RCA sequence-specific DNA binding [IEA] protein dimerization activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGQTETLSS TSNIPIAKAE PEQSADFSAS HKKRGPVSDR SSRRTSSEEV DLMPNVDDEV 60
61 DGDVKPKKIG RKNSDQEPSS KRKAQNRAAQ RAFRKRKEDH LKALETQVVT LKELHSSTTL 120
121 ENDQLRQKVR QLEEELRILK DGSFTFEMSL PHRNPSLSSL PTTGFSSNFA HMKDGISPQS 180
181 NLHLSPNSIE KPNMHQNVLH NDRSADNLNH RYQVPPTLVD SNSAQGTLSP ETPSSSDSPS 240
241 NLYLNYPKRK SITHLHHDCS ALSNGENGED VADGKQFCQK LSTACGSIAC SMLTKTTPHR 300
301 ASVDILSNLH ESTVSPPMAD ESVQRSSEVS KSIPNVELSL NVNQQFVSPF GGTDSFPLPT 360
361 DTGLDSLFEP DSAIENSHLK NVVMEPELFQ AWREPAESLD KEFFNDEGEI DDVFHNYFHN 420
421 SNENGDLITN SLHGLDFLEN ANESFPEQMY PFIKHNKDYI SNHPDEVPPD GLPQKGKHDT 480
481 SSQMPSENEI VPAKERAYLS CPKVWSKIIN HPRFESFDID DLCSKLKNKA KCSSSGVLLD 540
541 ERDVEAALNQ FN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
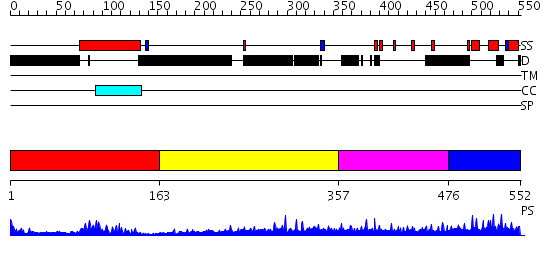
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | 9.0 | PAP1 | |
| 2 | View Details | [163..356] | 8.138992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [357..475] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [476..552] | 23.39794 | Solution structure of the oxidized form of the Yap1 redox domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)