













| General Information: |
|
| Name(s) found: |
X11L-PA /
FBpp0074225
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1167 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
plasma membrane [IDA] |
| Biological Process: |
synaptic vesicle targeting
[NAS]
synaptic vesicle docking during exocytosis [NAS] establishment or maintenance of cell polarity [NAS] neurotransmitter secretion [NAS] |
| Molecular Function: |
protein binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSNSCSDSS NESMVNPPHI QMNFGQQAQQ QQSHQHQQQQ QQQERYVWEM RTRSPNVTPA 60
61 STVLNSPDLN GELSYVPLQG LSPSGGGVQG SGSYGSNAGR DSGSYYYQRP QLHQIRLGRS 120
121 PGSQFETRDV LPQGLASPGS PTDVGKADGQ CLRDGEIVVF DDIDTNWMNK ASSGQRPGSN 180
181 EDVLKVTKMI GQLPIAEYEG SPRRFGSQPN TNTITMGGPR KRPPGFPQRV SPTTSVANNA 240
241 TAAAKASVGN LIDLVDHEEE RSGTLREKTP TFDYLYEFSE TRKVLEEFFK ANPEDEKRYT 300
301 DYTTESGDDV ASSPQEYPAT PMEQAYIGQR LARIPKDELY MVHRSPTKKP PSDQNPSTAY 360
361 QEHNDIELYI DSNSRSSGDL ADTELEANLR RHSRNFTLSP ETTDYDSNCG DLDSLSNDIN 420
421 CPTDFGKLYT SMPVLEDGLS SGHASDTENN VSVVCDKQQS SQSQPVHEHN GNGERLENDY 480
481 NVMSASLATL TTDASQSALI SDFASLPMPP APPPSSPPQI EIADASESMT MVPDADNPLD 540
541 SHYGAVYAAA LGGTHTEKSA GSGLGVTPVT AGGSLPSQAA SEIQEAMKEI RSALQRAKTQ 600
601 PEKLKFCDEV LPTDPDSPVW VPRKGTTIST AQGAAADEEP DTDLETDRLL GQQRHDEQDF 660
661 FADQSLADNN ANEGTDIASN GHLNGTSDNN NPNPMNNSKP QTYSTATIRQ GIGTSLTPNS 720
721 PDICQIVGTT DISISSPEKL QFTKSPTGSI KSLKDSANSD KKAKSRNKEG LLEPKVLIEG 780
781 VLFRARYLGS TQLVCEGQPT KSTRMMQAEE AVSRIKAPEG ESQPSTEVDL FISTEKIMVL 840
841 NTDLKEIMMD HALRTISYIA DIGDLVVLMA RRRFVPNSVV DPSITSPLGD VPTPGIGEEE 900
901 SPPKEPLSKH NRTPKMICHV FESDEAQFIA QSIGQAFQVA YMEFLKANGI ENESLAKEMD 960
961 YQEVLNSQEI FGDELEIFAK KELQKEVVVP KAKGEILGVV IVESGWGSML PTVVIANLMS1020
1021 SGAAARCGQL NIGDQLIAIN GMSLVGLPLS TCQSYIRNAK NQTAVKFTVV PCPPVVEVKI1080
1081 LRPKALFQLG FSVQNGVICS LLRGGIAERG GVRVGHRIIE INNQSVVAVP HDTIVKLLSS1140
1141 SVGEILMKTM PTSMFRLLTG QETPIYI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
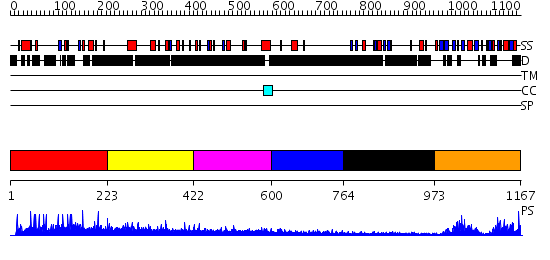
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..222] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [223..421] | 1.199994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [422..599] | 3.252997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [600..763] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [764..972] | 2.32 | X11 | |
| 6 | View Details | [973..1167] | 51.39794 | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)