













| General Information: |
|
| Name(s) found: |
mRNA-capping-enzyme-PA /
FBpp0073739
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 649 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
mRNA capping
[ISS]
protein amino acid dephosphorylation [IEA][NAS] |
| Molecular Function: |
protein tyrosine phosphatase activity
[IEA]
mRNA guanylyltransferase activity [ISS] polynucleotide 5'-phosphatase activity [IEA] protein tyrosine/serine/threonine phosphatase activity [IEA][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALTNRDRER GSGPLPNRWL YCPRKSDTII AERFLAFKTP LSNNFHDKMP IECTFQPEML 60
61 FEYCKTLKVK LGLWVDLTNT KRFYDRSAVE ELGAKYIKLQ CRGHGETPSP EQTHSFIEIV 120
121 DNFINERPFD VIAVHCTHGF NRTGFLIVCY LVERLDCSVS AALAIFASAR PPGIYKQDYI 180
181 NELYKRYEDT NAAPAAPEQP NWCLDYDDGN GDGFVQDNSS STSQQAGEQD DDAEEVEGED 240
241 AGGDCDASTS DGQPRKKRRR EMIIKNATFM AGVPGVRQVS DQPRLGDLQR KVQDWCDWKK 300
301 NGFPGAQPVS MDRENIKRLS EIPYRVSWKA DGTRYMMLID GRDEVYFFDR NHSCFQVENV 360
361 TFVESRNLNE HLDGTLVDGE MVLDKIGETV TPRYLIYDIV RLSNRDVRDE PFYPNRLDYI 420
421 KTEVIGPRIL GMKHGIINQR LQAFSVRGKD FWDIWMSARL LGEKFSRTLA HEPDGLIFQP 480
481 SKQPYTAGTC SDVFKWKPHE LNSVDFRLKI ITERGEGLLT KKVGFLYVGG HDAPYGRMQR 540
541 LTKETRELDN RIVECTMNQF GNWDFMRERT DKKNPNSYNT ARSVVDSIKH PVTKEYLLNF 600
601 IASSGYRNDH VMMPPPINSH NQNHNHNHFY PHGHGQHQRR PCVNPTLGN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
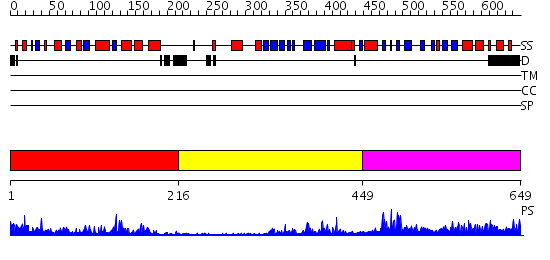
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | 32.39794 | CRYSTAL STRUCTURE OF THE HUMAN RNA guanylyltransferase and 5'- phosphatase | |
| 2 | View Details | [216..448] | 4.14 | No description for 2hivA was found. | |
| 3 | View Details | [449..649] | 37.154902 | mRNA capping enzyme alpha subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)