













| General Information: |
|
| Name(s) found: |
lz-PA /
FBpp0071255
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 826 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC][ISS]
|
| Biological Process: |
embryonic crystal cell differentiation
[IEP][IMP][TAS]
larval lymph gland hemocyte differentiation [TAS] embryonic hemopoiesis [IMP] hemopoiesis [TAS] embryonic hemocyte differentiation [IMP] positive regulation of transcription from RNA polymerase II promoter [IMP][TAS] regulation of transcription [IEA] compound eye cone cell differentiation [IMP] negative regulation of transcription from RNA polymerase II promoter [TAS] wound healing [IMP] cell fate determination [NAS] scab formation [IMP] spermathecum morphogenesis [TAS] signal transduction [NAS] regulation of transcription, DNA-dependent [IEA] lymph gland crystal cell differentiation [TAS] regulation of apoptosis [IMP] antennal development [TAS] defense response [NAS] compound eye development [TAS] crystal cell differentiation [IMP][NAS][TAS] eye morphogenesis [IMP] oogenesis [TAS] sensory organ precursor cell fate determination [TAS] cell fate commitment [TAS] positive regulation of compound eye retinal cell programmed cell death [IMP] cell differentiation [TAS] |
| Molecular Function: |
ATP binding
[IEA]
DNA binding [IDA] transcription factor activity [IEA][NAS] RNA polymerase II transcription factor activity [IGI][ISS] RNA polymerase II transcription factor activity, enhancer binding [NAS] sequence-specific DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHLHLLAAEK TPPSPSPNPT PTPSASPSGH TGAAGESQLD LASQTESQLQ LGLPLASGYG 60
61 LGLGLGLGLG LGLGQELVAD HSTTVAPVSV AGPGRLGRSI NGSGGSHHHH HLHHHYSPYH 120
121 HAHPYHPPHP HAPHHHHHHH PPYPYPPAGP HPPAMVTSSS TSPTGNGWSS STGDFKGITA 180
181 VATATGGGVG GATQGATAST GATAAEVLAV SSSASVGSSS PTGGASNGTA HSGHSGHTGG 240
241 HSSSTASNNN NNGASNSNSN NNNAVHQDLL WMERLVQKRQ QEHPGELVRT SNPYFLCSAL 300
301 PAHWRSNKTL PMAFKVVALA EVGDGTYVTI RAGNDENCCA ELRNFTTQMK NDVAKFNDLR 360
361 FVGRSGRGKS FTLTITVATS PPQVATYAKA IKVTVDGPRE PRSKTSPTGG PHYRALGLGQ 420
421 RPYIDGFPST KALHELESLR RSAKVAAVTT AAAAAATAAS AANAVAAAAA AVAVTPTGGG 480
481 GGVAAGGVAG GAGAGLVQQL SSNYSSPNST INSDCQVYKP NAPHIQAAEM MGAGEWTNGS 540
541 SSSAAAYYHS HAHHPHAHHA HAHLQHQMAL PPPPPPPAAA PVSVGVGGNG ATMGMGMGVG 600
601 VGMGMNHYGG GYDSANSLEA GQYAAHLPAV LPEMHGHGFA TDPYQTAGYG GGNTGGGSAS 660
661 KSELDYGGSY NQAWSNGYQN YQYGSCLATA QYGPQAAPPP QPPPPPPVVL CPQLYSTVNQ 720
721 NQIHLHLHSS EKLEQYLGTA TSADHLTIGS LTGSSRSSIE IGQDQYHQQV HHAQQQQQQQ 780
781 QQQQQVHHPQ QQQVESAGEV GGSGAGGVES AREEDVGDLS QVWRPY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
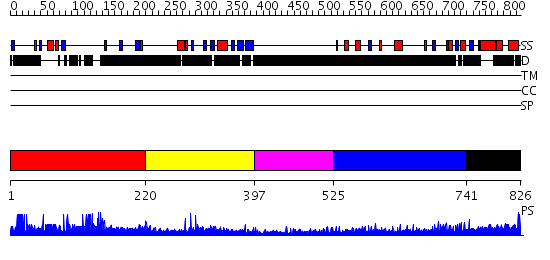
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..219] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [220..396] | 46.39794 | Acute myeloid leukemia 1 protein (AML1), RUNT domain | |
| 3 | View Details | [397..524] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [525..740] | 5.211981 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [741..826] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|