













| General Information: |
|
| Name(s) found: |
CG7033-PA /
FBpp0071226
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 535 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA]
chaperonin-containing T-complex [ISS] |
| Biological Process: |
mitotic spindle organization
[IMP]
protein folding [ISS] |
| Molecular Function: |
ATP binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEMSLNPVRV LKNEAQEEKA EMARLSSFIG AIAIGDLVKS TLGPKGMDKI LVATGRNAGH 60
61 VEVTNDGATI LRAVGVDNPA AKILVDMSRV QDEEVGDGTT SVTVLASELL REAEKLVEQK 120
121 LHPQIIVSGW RQATQVALEA LTAAAQDNSS SDEKFRNDLL NIARTTLSSK ILHQHKDFFA 180
181 NLAVDAVMRL KGSGELKSIQ IIKKSGGTLG DSFLDEGFLL DKKPGVHQPQ RIENARILIA 240
241 NTPMDTDKIK VFGSSIKVDS LAKIADLEMA EKEKMKEKVD KILKHKCNVF INRQLIYNYP 300
301 EQLFADARVM SIEHADFDGI ERLAYCTGGE IVSTFENPSL VKLGECDVIE QVMIGEDTLL 360
361 RFSGVKLGEA CTIVIRGATQ QILDEADRSL HDALCVLAAT VKESRIIFGG GCSEALMATA 420
421 VLKKAAETPG KEAIAIEAFA RALLSLPTAI ADNAGYDSAQ LVSELRAGHA QGKQTLGLDM 480
481 ELGKVADVRE LGITESFAVK RQVLMSASEA AEMILRVDNI IRCAPRRRVP DRGYC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
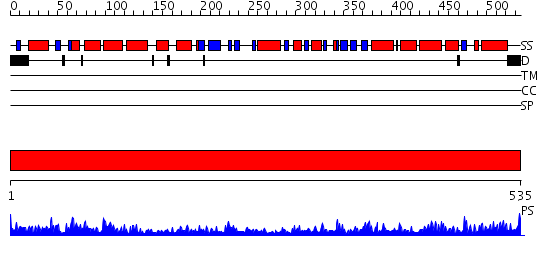
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..535] | 142.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)