













| General Information: |
|
| Name(s) found: |
CG7033-PC /
FBpp0071228
[FlyBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 533 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA]
chaperonin-containing T-complex [ISS] |
| Biological Process: |
mitotic spindle organization
[IMP]
protein folding [ISS] |
| Molecular Function: |
ATP binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLNPVRVLK NEAQEEKAEM ARLSSFIGAI AIGDLVKSTL GPKGMDKILV ATGRNAGHVE 60
61 VTNDGATILR AVGVDNPAAK ILVDMSRVQD EEVGDGTTSV TVLASELLRE AEKLVEQKLH 120
121 PQIIVSGWRQ ATQVALEALT AAAQDNSSSD EKFRNDLLNI ARTTLSSKIL HQHKDFFANL 180
181 AVDAVMRLKG SGELKSIQII KKSGGTLGDS FLDEGFLLDK KPGVHQPQRI ENARILIANT 240
241 PMDTDKIKVF GSSIKVDSLA KIADLEMAEK EKMKEKVDKI LKHKCNVFIN RQLIYNYPEQ 300
301 LFADARVMSI EHADFDGIER LAYCTGGEIV STFENPSLVK LGECDVIEQV MIGEDTLLRF 360
361 SGVKLGEACT IVIRGATQQI LDEADRSLHD ALCVLAATVK ESRIIFGGGC SEALMATAVL 420
421 KKAAETPGKE AIAIEAFARA LLSLPTAIAD NAGYDSAQLV SELRAGHAQG KQTLGLDMEL 480
481 GKVADVRELG ITESFAVKRQ VLMSASEAAE MILRVDNIIR CAPRRRVPDR GYC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
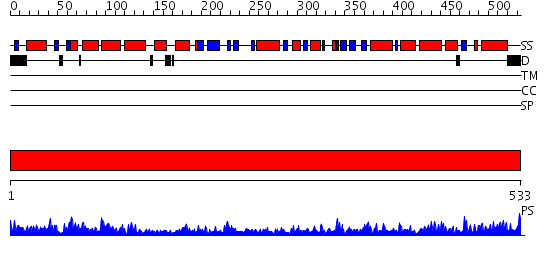
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..533] | 142.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)