













| General Information: |
|
| Name(s) found: |
Lim1-PA /
FBpp0071216
[FlyBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 505 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
cellular_component [ND] |
| Biological Process: |
biological_process
[ND]
imaginal disc-derived leg morphogenesis [IMP] negative regulation of transcription [IGI] regulation of transcription, DNA-dependent [IEA][ISS] positive regulation of transcription [IMP] regulation of transcription [IEA] leg disc proximal/distal pattern formation [IMP] |
| Molecular Function: |
transcription factor activity
[ISS]
zinc ion binding [IEA] sequence-specific DNA binding [IEA] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFGAVLHTS QHNIGRSEPP VGVGDPCAGC NKPILDKFLL NVLERAWHAS CVRCCECLQP 60
61 LTDKCFSRES KLYCRNDFFR RYGTKCSGCG QGIAPSDLVR KPRDKVFHLN CFTCCICRKQ 120
121 LSTGEQLYVL DDNKFICKDD YLLGKAPSCG HNSLSDSLMG SASEDDDDDD PPHLRATALG 180
181 LGVLGPNGPD SAGGPLGTSD ISVQSMSTDS KNTHDDSDQG SLDGDPDGRG DSQAENKSPD 240
241 DANGSKRRGP RTTIKAKQLE VLKTAFNQTP KPTRHIREQL AKETGLPMRV IQVWFQNKRS 300
301 KERRMKQITS MGRPPFFGGA RKMRGFPMNL SPGGLDDGPG FPYFAADAKF EFGYGGPFHP 360
361 HDGPYFPGHP GGPMPFNAPG GPMDHSGPIP MVNEFGLTPE ATFLGQAGGP PMQLQSAVGP 420
421 PPPPPPPSAE HLMAAAAAAA QQQAAQQQQQ QQQQQQQQQQ QNAQQAANQT QQQQQQNGPR 480
481 TNSPEFMSSA NFSEPQNMQN EGLVW |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
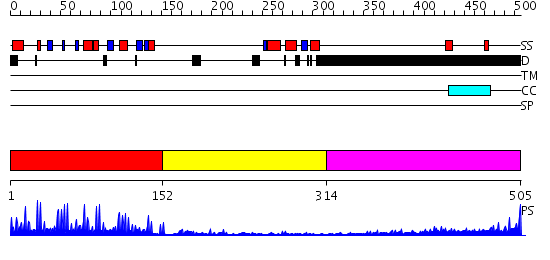
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 30.0 | No description for 2dfyX was found. | |
| 2 | View Details | [152..313] | 4.17 | TERNARY COMPLEX OF THE DNA BINDING DOMAINS OF THE OCT1 AND SOX2 TRANSCRIPTION FACTORS WITH A 19MER OLIGONUCLEOTIDE FROM THE HOXB1 REGULATORY ELEMENT | |
| 3 | View Details | [314..505] | 3.69897 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)