













| General Information: |
|
| Name(s) found: |
FBpp0311816
[FlyBase]
Ubi-p5E-PA / FBpp0070894 [FlyBase] |
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 534 amino acids |
Gene Ontology: |
|
| Cellular Component: |
polytene chromosome band
[IDA]
|
| Biological Process: |
protein modification process
[ISS]
ubiquitin-dependent protein catabolic process [ISS] |
| Molecular Function: |
protein binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGKQL EDGRTLSDYN 60
61 IQKESTLHLV LRLRGGMQIF VKTLTGKTIT LEVEPSDTIE NVKAKIQDKE GIPPDQQRLI 120
121 FAGKQLEDGR TLSDYNIQKE STLHLVLRLR GGMQIFVKTL TGKTITLEVE PSDTIENVKA 180
181 KIQDKEGIPP DQQRLIFAGK QLEDGRTLSD YNIQKESTLH LVLRLRGGMQ IFVKTLTGKT 240
241 ITLEVEPSDT IENVKAKIQD KEGIPPDQQR LIFAGKQLED GRTLSDYNIQ KESTLHLVLR 300
301 LRGGMQIFVK TLTGKTITLE VEPSDTIENV KAKIQDKEGI PPDQQRLIFA GKQLEDGRTL 360
361 SDYNIQKEST LHLVLRLRGG MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ 420
421 QRLIFAGKQL EDGRTLSDYN IQKESTLHLV LRLRGGMQIF VKTLTGKTIT LEVEPSDTIE 480
481 NVKAKIQDKE GIPPDQQRLI FAGKQLEDGR TLSDYNIQKE STLHLVLRLR GGAI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
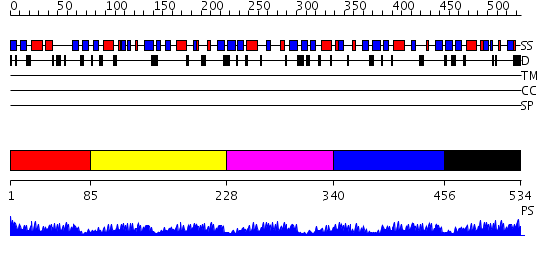
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 30.09691 | Solution Structure of S5a UIM-1/Ubiquitin Complex | |
| 2 | View Details | [85..227] | 4.84 | Crystal Structure of ISG15, the Interferon-Induced Ubiquitin Cross Reactive Protein | |
| 3 | View Details | [228..339] | 4.30103 | Elongin B | |
| 4 | View Details | [340..455] | 13.0 | Solution structure of Ubiquitin-like domain in SF3a120 | |
| 5 | View Details | [456..534] | 28.0 | Crystal structure of a multiple hydrophobic core mutant of ubiquitin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)