













| General Information: |
|
| Name(s) found: |
ovo-PE /
FBpp0291128
[FlyBase]
ovo-PB / FBpp0070706 [FlyBase] |
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1351 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC][ISS][NAS]
|
| Biological Process: |
adult feeding behavior
[IMP]
positive regulation of transcription from RNA polymerase II promoter [IMP] regulation of transcription [TAS] cuticle development [TAS] pheromone metabolic process [IMP] negative regulation of transcription from RNA polymerase II promoter [IMP] cuticle pigmentation [IMP] epidermal cell differentiation [IMP] oogenesis [TAS] female germ-line sex determination [NAS] germ-line sex determination [IGI][IMP] non-sensory hair organization [IMP][TAS] cuticle pattern formation [IMP] |
| Molecular Function: |
DNA binding
[IDA][TAS]
zinc ion binding [IEA] RNA polymerase II transcription factor activity [ISS] sequence-specific DNA binding [IDA] transcription activator activity [IMP] transcription repressor activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPKIFLIKNR LHQQQQRLLE SQNLLQHKNQ DDERLVPPLS PSGSGSGPSP TPTSQPPPEP 60
61 QGQGQQVLGQ VPDSDQQPLS LTRKRFHHRR HYFGQSRHSL DHLNQNQSPN PNANANPNQI 120
121 QNPAELEVEC ATGQVQNENF AAELLQRLTP NTATTAQNNI VNNLVNNSRA ATSVLATKDC 180
181 TIENSPISIP KNQRAEDEEE QEDQEKEKPA EREREKSDER TEQVEKEERV EREEEEDDEV 240
241 DVGVEAPRPR FYNTGVVLTQ AQRKEYPQEP KDLSLTIAKS SPASPHIHSD SESDSDSDGG 300
301 CKLIVDEKPP LPVIKPLSLR LRSTPPPADQ RPSPPPPRDP APAVRCSVIQ RAPQSQLPTS 360
361 RAGFLLPPLD QLGPEQQEPI DYHVPKRRSP SYDSDEELNA RRLERARQVR EARRRSTILA 420
421 ARVLLAQSQR LNPRLVRSLP GILAAAAGHG RNSSSSSGAA GQGFQSSGFG SQNSGSGSSS 480
481 GNQNAGSGAG SPGSGAGGGG GMGGGRDGRG NYGPNSPPTG ALPPFYESLK SGQQSTASNN 540
541 TGQSPGANHS HFNANPANFL QNAAAAAYIM SAGSGGGGCT GNGGGGASGP GGGPSANSGG 600
601 GGGGGGGNGY INCGGVGGPN NSLDGNNLLN FASVSNYNES NSKFHNHHHH HQHNNNNNNN 660
661 GGQTSMMGHP FYGGNPSAYG IILKDEPDIE YDEAKIDIGT FAQNIIQATM GSSGQFNASA 720
721 YEDAIMSDLA SSGQCPNGAV DPLQFTATLM LSSQTDHLLE QLSDAVDLSS FLQRSCVDDE 780
781 ESTSPRQDFE LVSTPSLTPD SVTPVEQHNT NTTQLDVLHE NLLTQLTHNI VRGGSNQQQQ 840
841 HHQQHGVQQQ QQQQHSVQQQ QQHNVQQQHG VQQQHVQQQP PPSYQHATRG LMMQQQPQHG 900
901 GYQQQAAIMS QQQQQLLSQQ QQQSHHQQQQ QQQHAAAYQQ HNIYAQQQQQ QQQQHHQQQQ 960
961 QQQHHHFHHQ QQQQPQPQSH HSHHHGHGHD NSNMSLPSPT AAAAAAAAAA AAAAAAAAHL1020
1021 QRPMSSSSSS GGTNSSNSSG GSSNSPLLDA NAAAAAAAAL LDTKPLIQSL GLPPDLQLEF1080
1081 VNGGHGIKNP LAVENAHGGH HRIRNIDCID DLSKHGHHSQ HQQQQGSPQQ QNMQQSVQQQ1140
1141 SVQQQQSLQQ QQQQQHHQHH SNSSASSNAS SHGSAEALCM GSSGGANEDS SSGNNKFVCR1200
1201 VCMKTFSLQR LLNRHMKCHS DIKRYLCTFC GKGFNDTFDL KRHTRTHTGV RPYKCNLCEK1260
1261 SFTQRCSLES HCQKVHSVQH QYAYKERRAK MYVCEECGHT TCEPEVHYLH LKNNHPFSPA1320
1321 LLKFYDKRHF KFTNSQFANN LLGQLPMPVH N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
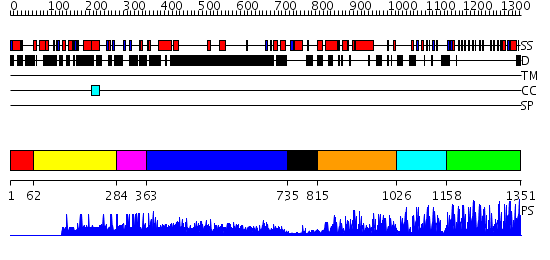
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..61] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [62..283] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [284..362] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [363..734] | 43.30103 | No description for 1y0fA was found. | |
| 5 | View Details | [735..814] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [815..1025] | 1.513997 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1026..1157] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1158..1351] | 67.69897 | No description for 2i13A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.52 |
Source: Reynolds et al. (2008)