













| General Information: |
|
| Name(s) found: |
ph-p-PA /
FBpp0070416
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
PRC1 complex [TAS] nucleolus [IDA] |
| Biological Process: |
gene silencing
[IGI]
ovarian follicle cell stalk formation [IMP] response to ecdysone [IMP] syncytial blastoderm mitotic cell cycle [IMP] germarium-derived female germ-line cyst encapsulation [IMP] chromatin silencing [NAS] central nervous system neuron development [IMP] |
| Molecular Function: |
DNA binding
[NAS]
zinc ion binding [IEA] protein binding [IPI] chaperone binding [IDA] chromatin binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDRRALKFMQ KRADTESDTT TPVSTTASQG ISASAILAGG TLPLKDNSNI REKPLHHNYN 60
61 HNNNNSSQHS HSHQQQQQQQ VGGKQLERPL KCLETLAQKA GITFDEKYDV ASPPHPGIAQ 120
121 QQATSGTGPA TGSGSVTPTS HRHGTPPTGR RQTHTPSTPN RPSAPSTPNT NCNSIARHTS 180
181 LTLEKAQNPG QQVAATTTVP LQISPEQLQQ FYASNPYAIQ VKQEFPTHTT SGSGTELKHA 240
241 TNIMEVQQQL QLQQQLSEAN GGGAASAGAG GAASPANSQQ SQQQQHSTAI STMSPMQLAA 300
301 ATGGVGGDWT QGRTVQLMQP STSFLYPQMI VSGNLLHPGG LGQQPIQVIT AGKPFQGNGP 360
361 QMLTTTTQNA KQMIGGQAGF AGGNYATCIP TNHNQSPQTV LFSPMNVISP QQQQNLLQSM 420
421 AAAAQQQQLT QQQQQFNQQQ QQQLTQQQQQ LTAALAKVGV DAQGKLAQKV VQKVTTTSSA 480
481 VQAATGPGST GSTQTQQVQQ VQQQQQQTTQ TTQQCVQVSQ STLPVGVGGQ SVQTAQLLNA 540
541 GQAQQMQIPW FLQNAAGLQP FGPNQIILRN QPDGTQGMFI QQQPATQTLQ TQQNQIIQCN 600
601 VTQTPTKART QLDALAPKQQ QQQQQVGTTN QTQQQQLAVA TAQLQQQQQQ LTAAALQRPG 660
661 APVMPHNGTQ VRPASSVSTQ TAQNQSLLKA KMRNKQQPVR PALATLKTEI GQVAGQNKVV 720
721 GHLTTVQQQQ QATNLQQVVN AAGNKMVVMS TTGTPITLQN GQTLHAATAA GVDKQQQQLQ 780
781 LFQKQQILQQ QQMLQQQIAA IQMQQQQAAV QAQQQQQQQV SQQQQVNAQQ QQAVAQQQQA 840
841 VAQAQQQQRE QQQQVAQAQA QHQQALANAT QQILQVAPNQ FITSHQQQQQ QQLHNQLIQQ 900
901 QLQQQAQAQV QAQVQAQAQQ QQQQREQQQN IIQQIVVQQS GATSQQTSQQ QQHHQSGQLQ 960
961 LSSVPFSVSS STTPAGIATS SALQAALSAS GAIFQTAKPG TCSSSSPTSS VVTITNQSST1020
1021 PLVTSSTVAS IQQAQTQSAQ VHQHQQLISA TIAGGTQQQP QGPPSLTPTT NPILAMTSMM1080
1081 NATVGHLSTA PPVTVSVTST AVTSSPGQLV LLSTASSGGG GSIPATPTKE TPSKGPTATL1140
1141 VPIGSPKTPV SGKDTCTTPK SSTPATVSAS VEASSSTGEA LSNGDASDRS STPSKGATTP1200
1201 TSKQSNAAVQ PPSSTTPNSV SGKEEPKLAT CGSLTSATST STTTTITNGI GVARTTASTA1260
1261 VSTASTTTTS SGTFITSCTS TTTTTTSSIS NGSKDLPKAM IKPNVLTHVI DGFIIQEANE1320
1321 PFPVTRQRYA DKDVSDEPPK KKATMQEDIK LSGIASAPGS DMVACEQCGK MEHKAKLKRK1380
1381 RYCSPGCSRQ AKNGIGGVGS GETNGLGTGG IVGVDAMALV DRLDEAMAEE KMQTEATPKL1440
1441 SESFPILGAS TEVPPMSLPV QAAISAPSPL AMPLGSPLSV ALPTLAPLSV VTSGAAPKSS1500
1501 EVNGTDRPPI SSWSVDDVSN FIRELPGCQD YVDDFIQQEI DGQALLLLKE KHLVNAMGMK1560
1561 LGPALKIVAK VESIKEVPPP GEAKDPGAQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
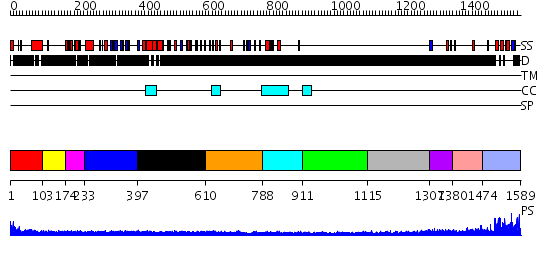
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [103..173] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [174..232] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [233..396] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [397..609] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [610..787] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [788..910] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [911..1114] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1115..1306] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1307..1379] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1380..1473] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1474..1589] | 31.0 | Hetero SAM domain structure of Ph and Scm. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. | |||||||||
| 8 | No functions predicted. | |||||||||
| 9 | No functions predicted. | |||||||||
| 10 | No functions predicted. | |||||||||
| 11 | No functions predicted. | |||||||||
| 12 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)