













| General Information: |
|
| Name(s) found: |
deltaCOP-PA /
FBpp0070355
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 532 amino acids |
Gene Ontology: |
|
| Cellular Component: |
clathrin adaptor complex
[IEA]
COPI vesicle coat [ISS][NAS] |
| Biological Process: |
phagocytosis, engulfment
[IMP]
retrograde vesicle-mediated transport, Golgi to ER [ISS] intracellular protein transport [IEA] |
| Molecular Function: |
protein binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLIAAAVCT KNGKVILSRQ FVEMTKARIE GLLAAFPKLM TAGKQHTYVE TDSVRYVYQP 60
61 MEKLYMLLIT TKASNILEDL ETLRLFSKVI PEYSHSLDEK EIVENAFNLI FAFDEIVALG 120
121 YRESVNLAQI KTFVEMDSHE EKVYQAVRQT QERDARQKMR EKAKELQRQR MEASKRGGPS 180
181 LGGIGSRSGG FSADGIGSSG VSSSSGASSA NTGITSIDVD TKSKAAASKP ASRNALKLGG 240
241 KSKDVDSFVD QLKNEGEKIA NLAPAAPAGG SSAAASASAA AKAAIASDIH KESVHLKIED 300
301 KLVVRLGRDG GVQQFENSGL LTLRITDEAY GRILLKLSPN HTQGLQLQTH PNVDKELFKS 360
361 RTTIGLKNLG KPFPLNTDVG VLKWRFVSQD ESAVPLTINC WPSDNGEGGC DVNIEYELEA 420
421 QQLELQDVAI VIPLPMNVQP SVAEYDGTYN YDSRKHVLQW HIPIIDAANK SGSMEFSCSA 480
481 SIPGDFFPLQ VSFVSKTPYA GVVAQDVVQV DSEAAVKYSS ESILFVEKYE IV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
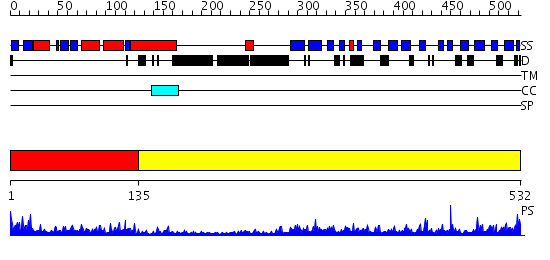
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 37.09691 | AP1 clathrin adaptor core | |
| 2 | View Details | [135..532] | 65.0 | Second domain of Mu2 adaptin subunit (ap50) of ap2 adaptor; Mu2 adaptin (clathrin coat assembly protein AP50) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)