













| General Information: |
|
| Name(s) found: |
FBpp0298002
[FlyBase]
elav-PA / FBpp0070086 [FlyBase] |
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 483 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][TAS]
Cajal body [IDA] |
| Biological Process: |
RNA metabolic process
[NAS]
central nervous system development [IMP] nervous system development [NAS] negative regulation of mRNA 3'-end processing [IDA] |
| Molecular Function: |
nucleotide binding
[IEA]
RNA binding [IDA] mRNA binding [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFIMANTGA GGGVDTQAQL MQSAAAAAAV AATNAAAAPV QNAAAVAAAA QLQQQQVQQA 60
61 ILQVQQQQTQ QAVAAAAAAV TQQLQQQQQA VVAQQAVVQQ QQQQAAAVVQ QAAVQQAVVP 120
121 QPQQAQPNTN GNAGSGSQNG SNGSTETRTN LIVNYLPQTM TEDEIRSLFS SVGEIESVKL 180
181 IRDKSQVYID PLNPQAPSKG QSLGYGFVNY VRPQDAEQAV NVLNGLRLQN KTIKVSFARP 240
241 SSDAIKGANL YVSGLPKTMT QQELEAIFAP FGAIITSRIL QNAGNDTQTK GVGFIRFDKR 300
301 EEATRAIIAL NGTTPSSCTD PIVVKFSNTP GSTSKIIQPQ LPAFLNPQLV RRIGGAMHTP 360
361 VNKGLARFSP MAGDMLDVML PNGLGAAAAA ATTLASGPGG AYPIFIYNLA PETEEAALWQ 420
421 LFGPFGAVQS VKIVKDPTTN QCKGYGFVSM TNYDEAAMAI RALNGYTMGN RVLQVSFKTN 480
481 KAK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
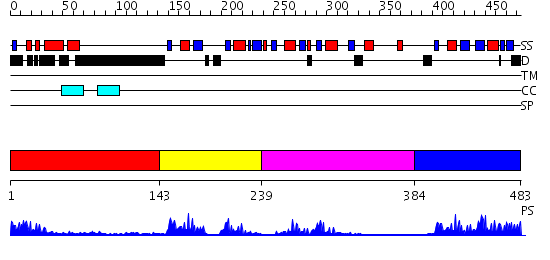
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | 1.42 | No description for 2j63A was found. | |
| 2 | View Details | [143..238] | 35.0 | Hu antigen D (Hud) | |
| 3 | View Details | [239..383] | 4.13 | CBP20, 20KDa nuclear cap-binding protein | |
| 4 | View Details | [384..483] | 21.522879 | Hu antigen C (Huc) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)