













| General Information: |
|
| Name(s) found: |
Sox100B-PA /
FBpp0085097
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 529 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
|
| Biological Process: |
male gonad development
[IDA][IMP]
regulation of transcription [ISS] |
| Molecular Function: |
transcription factor activity
[ISS]
DNA bending activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDSSSSNCS KDRAKPVETL VLANYALKAE QKKAQGQGGR KEDERITTAV MKVLEGYDWN 60
61 LVQASAKAPT DRKKEHIKRP MNAFMVWAQA ARRVMSKQYP HLQNSELSKS LGKLWKNLKD 120
121 SDKKPFMEFA EKLRMTHKQE HPDYKYQPRR KKARVLPSQQ SGEGGSPGPE MTLSATMGSS 180
181 GKPRSSNSNG QRRAGKGNAA ADLGSCASTI SHANVGSNSS DVFSNEAFMK SLNSACAASL 240
241 MEQSLIETGL DSPCSTASSM SSLTPPATPY NVAPSNAKAS AANNPSLLLR QLSEPVANAG 300
301 DGYGVLLEAG REYVAIGEVN YQGQSAGVQS GAEGGGAGQE MDFLENINGY GGYTGSRVSY 360
361 PAYSYPANGG HFATEEQQQQ QALQASEALN YKPAAADIDP KEIDQYFMDQ MLPMTQHHHP 420
421 HHTHPLHHPL HHSPPLNSSA SLSSACSSAS SQQPVAEYYE HLGYSPAASS ASQNPNFGPQ 480
481 QPYANGAASM TPTLGDPAPQ QELQSQQQEQ QHQNPSQHHL WGTYTYVNP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
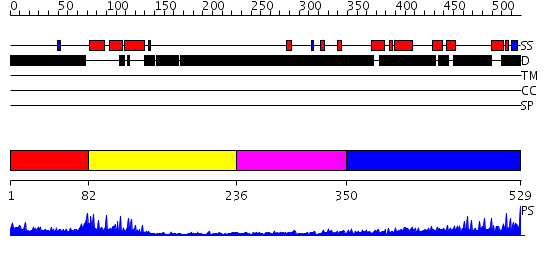
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | 12.221849 | NMR STRUCTURE OF RAT HMG1 HMGA FRAGMENT | |
| 2 | View Details | [82..235] | 26.0 | No description for 2gzkA was found. | |
| 3 | View Details | [236..349] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [350..529] | 13.213997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)