













| General Information: |
|
| Name(s) found: |
SNF4Agamma-PQ /
FBpp0290802
[FlyBase]
SNF4Agamma-PP / FBpp0290801 [FlyBase] SNF4Agamma-PK / FBpp0110239 [FlyBase] SNF4Agamma-PJ / FBpp0110238 [FlyBase] SNF4Agamma-PE / FBpp0083425 [FlyBase] SNF4Agamma-PC / FBpp0083424 [FlyBase] |
| Description(s) found:
Found 76 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 906 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
positive regulation of cell cycle
[IGI]
protein amino acid phosphorylation [ISS] cholesterol homeostasis [IMP] |
| Molecular Function: |
AMP-activated protein kinase activity
[ISS][NAS]
protein serine/threonine kinase activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHGITHLYRQ HAVEKQLSGG ESWSRQIYPS YGSSADASSQ GSRLYSVDSS SSSSSNSNNS 60
61 GGGGSGTAGV ENLGGVTGSA YAQWQSDRHS LPQAVPDAPR SLANHHYRNS PTHQAHYQQP 120
121 TSGSLKRTPS SKRSFLERSH SPAIYGSMRR SAGPDFGSPP PTAPVGGGGY FPHDLDDIYE 180
181 SQTDHQYFTH TGASGPQQQR PTAISIFHRV SAAHTSLDDY TLPGDASRQS MNSTDSGVSS 240
241 GPFNRQRFDN SSFAKSLSID EQDPQHSPHK EGHSKHGKHH HHHSHHHHSI HELVKHFGKK 300
301 MHLWPRKHHD AQSVCTSPQN DPQENFRTRS KSLDVNTLSR PNRILDDCGA TYKIYDRIVK 360
361 EGAHMRRASA DLEKRRASVG AAGRGLRGDG TLDPHHAAIL FRDSRGLPVA DPFLEKVNLS 420
421 DLEEDDSQIF VKFFRFHKCY DLIPTSAKLV VFDTQLLVKK AFYALVYNGV RAAPLWDSEK 480
481 QQFVGMLTIT DFIKILQMYY KSPNASMEQL EEHKLDTWRS VLHNQVMPLV SIGPDASLYD 540
541 AIKILIHSRI HRLPVIDPAT GNVLYILTHK RILRFLFLYI NELPKPAYMQ KSLRELKIGT 600
601 YNNIETADET TSIITALKKF VERRVSALPL VDSDGRLVDI YAKFDVINLA AEKTYNDLDV 660
661 SLRKANEHRN EWFEGVQKCN LDESLYTIME RIVRAEVHRL VVVDENRKVI GIISLSDILL 720
721 YLVLRPSGEG VGGSESSLRA SDPVLLRKVA EVEIPATAAA ATTTTPPRSP SAGSGNRSLI 780
781 EDIPEEETAP ARSDDADSDN NKSASEDKAN NNQHDQTTTA ATANGDSNNS PVEVSFADEA 840
841 QEEEAADQVE RSNCDDDDQP ALAEIERKNA SMDDDEDDGM SSAVSAASAL GQSLTPAAQE 900
901 MALVSE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
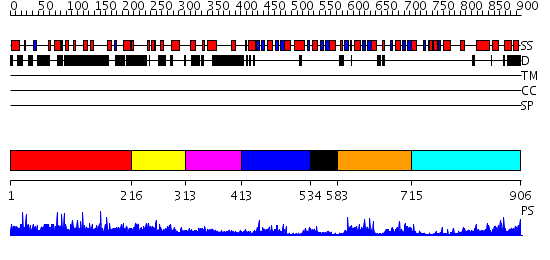
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [216..312] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [313..412] | 1.166991 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [413..533] | 51.39794 | No description for 2v8qE was found. | |
| 5 | View Details | [534..582] | 2.65 | Crystal structure of CBS domain-containing predicted protein (TM0935) from Thermotoga maritima at 1.87 A resolution | |
| 6 | View Details | [583..714] | 22.045757 | No description for 2uv4A was found. | |
| 7 | View Details | [715..906] | 19.522879 | Inosine monophosphate dehydrogenase (IMPDH) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)