













| General Information: |
|
| Name(s) found: |
Gyc-89Db-PA /
FBpp0082745
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 669 amino acids |
Gene Ontology: |
|
| Cellular Component: |
guanylate cyclase complex, soluble
[ISS]
|
| Biological Process: |
cGMP biosynthetic process
[IEA]
intracellular signaling pathway [IEA] |
| Molecular Function: |
guanylate cyclase activity
[IDA][ISS]
heme binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYGMLYESVQ HYIQQEYGME TWRKVCQIVD CKHQSFKTHQ IYPDKLMPDF AAALSASTGE 60
61 SFDFCMNFFG RCFVRFFSNF GYDKMIRSTG RYFCDFLQSI DNIHVQMRFT YPKMKSPSMQ 120
121 LTNMDDDGAV ILYRSGRTGM SKYLIGQMTE VAKEFYGLDM TAYVLESQND ICGGTAGPIK 180
181 LTEGPLTVIV KYRLDFDNRD YMAKRVNVIA HPSQLKMPSV DLNVFLELFP FTIVLDHDMK 240
241 ITLAGEKIVE TWILHNPGVN PKTFIGSHIL ERFKCRRPKD TQIQWETILQ MRTVLFEFEL 300
301 IRTGHNRAAY DAALNFDFEN FDEASSLNEA QAMALASAKE FSAENAKEEA AAAATSKDEI 360
361 DPATGQRRHS VGLRSILLKG QMFYIKDVDS LIFLCSPLIE NLDELHGIGL YLNDLNPHGL 420
421 SRELVMAGWQ HCSKLEIMFE KEEQRSDELE KSLELADSWK RQGDELLYSM IPRPIAERMR 480
481 KSEEHVCQSF EEVSVIFIEV MNIYDSGSNN IQDAMQAVTT LNKVFSALDE EIISPFVYKV 540
541 ETVGMVYMAV SGAPDVNPLH AEHACDLALR VMKKVKAHAL PGVAIRVGIN SGPVVAGVVG 600
601 MKVPRYCLFG DTVNTASRME SSSDPWMIQL SNYTALKVQK VGYKVEARGF VKVKGKGEME 660
661 TYWLLEGPE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
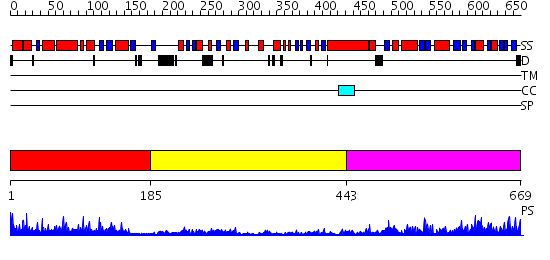
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | 42.39794 | No description for 2o09A was found. | |
| 2 | View Details | [185..442] | 3.69897 | Crystal structure of the phosphorylated Interleukin-2 tyrosine kinase catalytic domain | |
| 3 | View Details | [443..669] | 50.0 | Crystal structure of a mutant form of the mycobacterial adenylyl cyclase Rv1625c |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)