













| General Information: |
|
| Name(s) found: |
FBpp0311095
[FlyBase]
FBpp0311094 [FlyBase] Nsf2-PA / FBpp0082346 [FlyBase] |
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 752 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[ISS][NAS]
|
| Biological Process: |
ER to Golgi vesicle-mediated transport
[ISS][NAS]
synaptic vesicle priming [NAS] vesicle-mediated transport [NAS] neuromuscular synaptic transmission [IMP] regulation of synaptic growth at neuromuscular junction [IMP] intra-Golgi vesicle-mediated transport [NAS] neurotransmitter secretion [NAS] |
| Molecular Function: |
ATP binding
[IEA]
nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIEKAHRMR AIKCPTDELS LTNKAIVNVS DFTEEVKYVD ISPGPGLHYI FALEKISGPE 60
61 LPLGHVGFSL VQRKWATLSI NQEIDVRPYR FDASADIITL VSFETDFLQK KTTTQEPYDS 120
121 DEMAKEFLMQ FAGMPLTVGQ TLVFQFKDKK FLGLAVKTLE AVDPRTVGDS LPKTRNVRFG 180
181 RILGNTVVQF EKAENSVLNL QGRSKGKIVR QSIINPDWDF GKMGIGGLDK EFNAIFRRAF 240
241 ASRVFPPELV EQLGIKHVKG ILLYGPPGTG KTLMARQIGT MLNAREPKIV NGPQILDKYV 300
301 GESEANIRRL FAEAEEEEKR LGPNSGLHII IFDEIDAICK ARGSVAGNSG VHDTVVNQLL 360
361 AKIDGVEQLN NILVIGMTNR RDMIDEALLR PGRLEVQMEI SLPNEQGRVQ ILNIHTKRMR 420
421 DFNKIASDVD NNEIAAKTKN FSGAELEGLV RAAQSTAMNR LIKADSKVHV DPEAMEKLRV 480
481 TRADFLHALD NDIKPAFGAA QEMLENLLAR GIINWGPPVT ELLEDGMLSV QQAKATESSG 540
541 LVSVLIEGAP NSGKSALAAN LAQLSDFPFV KVCSPEDMVG FTESAKCLHI RKIFDDAYRS 600
601 TLSCIVVDNV ERLLDYGPIG PRYSNLTLQA LLVLLKKQPP KGRKLLILCT SSRRDVLEEM 660
661 EMLSAFTSVL HVSNLSTPEN VLAVLDDSDL FSPEELQSIA RKMAGKRLCI GIKKLLALID 720
721 MIRQSEPHQR VIKFLSKMEE EGGLEMDRVQ GH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
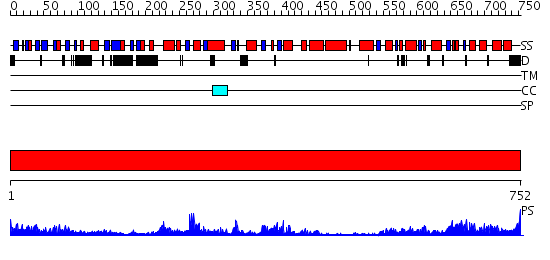
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..752] | 128.0 | VCP/p97 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)