













| General Information: |
|
| Name(s) found: |
FBpp0312064
[FlyBase]
FBpp0312063 [FlyBase] rin-PE / FBpp0082267 [FlyBase] rin-PB / FBpp0082265 [FlyBase] |
| Description(s) found:
Found 74 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 690 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA]
catalytic step 2 spliceosome [IDA] precatalytic spliceosome [IDA] cytosol [IDA] |
| Biological Process: |
compound eye photoreceptor fate commitment
[IMP]
ommatidial rotation [IMP] Ras protein signal transduction [IGI] transport [IEA] nuclear mRNA splicing, via spliceosome [IC] |
| Molecular Function: |
nucleotide binding
[IEA]
nucleic acid binding [IEA] protein binding [ISS] mRNA binding [ISS] SH3 domain binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVMDATQSQQ PSPQSVGREF VRQYYTLLNK APNHLHRFYN HNSSYIHGES KLVVGQREIH 60
61 NRIQQLNFND CHAKISQVDA QATLGNGVVV QVTGELSNDG QPMRRFTQTF VLAAQSPKKY 120
121 YVHNDIFRYQ DLYIEDEQDG ESRSENDEEH DVQVVGGTVD QAVQVAGDVV AQQPQQQQAP 180
181 PQPQAQVVVP QQVAPVAAGG AAPQQIFYPL PAGAAAGRPV AVLPPAPQAA AVPLPAQFTV 240
241 PPPQPIQNGV VSHEELQQQQ VLVPPSVSPL VQQQQPTGTP VLPIVPVPSA GFQQSPLVAP 300
301 QLIQQQQQQQ PAQLPPQQQQ QQQPIQQQQP LQQQQQPLAE PEEVEVAPRQ QKPPTPVEDF 360
361 KTIHEQQQQE KYEAAKQQQN EPKTYANLFK STSSSPSGFV NAALQQQQQL QQQQQQQQSI 420
421 SSNTYNSSSA TSGGGNGSQV SYSTTASSYN NSNGNSRLDN GGPLPQRNNS IRNNKGDFEQ 480
481 RRSSNTQQFG DNQQLFLGNI PHHASEDDLR EIFSRFGNVL ELRILSKAGN KVPPGMRSPL 540
541 NYGFITYDDP EAVQKCLANC PLYFPENSPD GQKLNVEEKK PRVRNDMVPR QQIGGNNLNN 600
601 NMGRLGGNNG PQSRPMGNNN GSGGGMMRNN AGGNNLRQGG GGGNGAPRLG GGGGFGQRQD 660
661 NRTGGGNNQS NGPLRGAGNG QSGGGNYGRR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
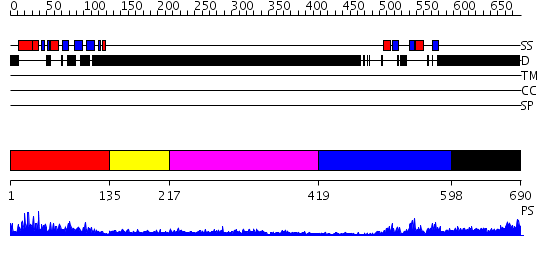
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 30.522879 | Nuclear transport factor-2 (NTF2) | |
| 2 | View Details | [135..216] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [217..418] | 2.235995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [419..597] | 33.221849 | Crystal Structure of UP1 Complexed With d(TTAGGGTTAG(6-MI)G); A Human Telomeric Repeat Containing 6-methyl-8-(2-deoxy-beta-ribofuranosyl)isoxanthopteridine (6-MI) | |
| 5 | View Details | [598..690] | 1.02 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)