













| General Information: |
|
| Name(s) found: |
FBpp0082058
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2016 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
|
| Biological Process: |
RNA splicing
[IMP]
transcription [IEA] regulation of transcription [ISS] |
| Molecular Function: |
transcription factor activity
[ISS]
zinc ion binding [IEA] protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSVFTEAP SARPAAAEAD SNLVVVYSKS GISFDERAIG NIMEEKSIST ISVVRITSPT 60
61 PSMVDQEEDE RLEELANFME TATDSELDSD NGSQSGGDNA SEMDEHDEEN HNTGDDNNSD 120
121 STETEADAPR TRKRVGRKPK AIKKRKRRKP KERPQIVGLR VEQFYADNTA DMVVKNALKL 180
181 AGVSVYKRTA ETDALMEVIR NDHNYTPFTS PEQLKNHKRD EKMAMEMQAQ SRKIIMQAPG 240
241 SVKLINAKKR VQAIPLSPVQ VKVQRLPVKP HQVQQAAQLQ PKVQLIRKPI HSPLPRQKPE 300
301 IIANNSVNAR LRPTRAPVKV IAPPVQEYIE DDDDAQSSDS AEEENADKAY DSEEMSEESD 360
361 EFQESDNDRD SDMDFDMRHS NRRNPNKRKR VRKVVRQTPS KPMKPLPPPP TTPQHFPELK 420
421 KRKEVLEPKA PQIGQIIQMP ATKSVPAVFA LGRGLPSTSF LKIRPTHQSL PVKRPPLPTT 480
481 PANSNVRRMP AVQLGRIVNS PQEVKEIIIN KNMASPKGVF TNLNTLLGED NNAATKSSPV 540
541 PLRHRAVMSP STPRSSYNQQ MSPIVVPVPA QAHTPSKGFM PIGVDTAQSH KLPAQIVIET 600
601 HQSSSELAAE NDKQLDLINS IVQDELLKST LVEQPVVNAD EDIPKLVKML ESTAADLDPP 660
661 PVSLPTNIFP APEMQGVGNA AAADPNNIMD TANEDEITAD FLQHVVGLIE EDKQFEAEVV 720
721 KQVLASTEPG TLDAIVSMPT SIEPVDVPQA HTNLLPNASL TEPAQSMTSL PIACSTPSRS 780
781 VAASTPPTSA KVVRGYGRVI YLPPIEAPTT RAKRRAQFPS APGMAATSSS DAGNLSFGES 840
841 SLDASINQPL NTSSLSNDSQ PGSGPKRPNP REPSMARRST APRRSKKLDA SQNNDPDASE 900
901 SQEDDDDPNK LWCICRQPHN NRFMICCDLC EDWFHGTCVG VTKAMGTDME NKGIDWKCPK 960
961 CVKRQEERSQ PRITDMLVTR PTTQPEQRPS ETKVLTTTAE IVQVAAPSAP RRTLPVVLTV1020
1021 ASSPMRIPMA KPAKKFPTGA ISHQQQQQLN FIRLGPSPGK RISETLCVVC KRPASTSSVY1080
1081 CGEECIRKYA QSAIQAHAAT KGPLPQNAGA QSLLNNSFDA KKNKKKDLFE DVLRQADTVS1140
1141 KVERINVFER KSGRVITGHM APSAHQFRKW LQENPSFEVL PSGTVQSADA EKRLLKGAPE1200
1201 AATSTSEPAV LGVAKKPPEG PAKLSHPQNT TVQASHQLGI SSVRPLAKKD KEKTTPTVQA1260
1261 PTPNRIAAGK PEPVRIGIRR SLREQLLARI KEAQAAEENS GQPTTQWPTV LEVDQFVKNV1320
1321 ELEMFNSFGR DVGAKYKAKY RSLMFNIKDR KNRTLFEKIC AKQVEPRQLV RMTPEQLASQ1380
1381 ELAKWREEEN RHQLDMIKKS ELDLLACAQN YVVKTHKGEE VIATKVDVTL PEENVSEASS1440
1441 MADTKQTSLV SEPSTSTLER STSREKAGSS KEKRHKSHKH HHRKRSRSRS NSRGRSVDKR1500
1501 HRRQHNEGGE QPGSGEREHR SREKQSRERD EIVPSPLPKK RDENDQSPVP KKTAEKKTEA1560
1561 SAYNLVDQIL ESEKTVEQAA NLGKPKPSPK PLPTLPSSLK APEPMDNYSR YLHGLTTSSL1620
1621 WSGTLKMIDL ADFEIVMYPV QGNCHQLGNL MPSQMDVIGR ITRVNVWEYI KKLKKSPTKE1680
1681 VVIVNIFPAS PSETYKFDLF FEYLDSRQRL GVLGVDSDQI RDFYIFPLGS GDKLPPALQT1740
1741 AEPVPFYEEA QRPNTLLGII VRCLSKRPAD AVPSTPSVPT PVPALSSKVA KRSRSSISYP1800
1801 AQSSPKRKIS THSTSSKDDE FDIDAIIKAP IAKLQKTAPK VVPQPSVPND ADEPYSPGGS1860
1861 DDELVPSAPQ RPNDLERQVN EINKQIAAQQ MEIAGLLKVE PTGSASSSNV LAAISIPANL1920
1921 SKILASIKDK TNLPSSADDG DEEYNPEDDI TATSSFASKK PRSKGRLAHL SEAELLSMVP1980
1981 DNLAGVIPRS SRTRHQLAQS PLTPTPLPPP PPPPGV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
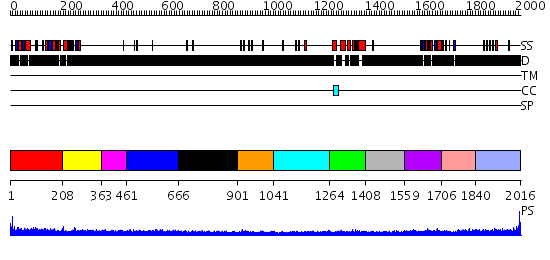
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..207] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [208..362] | 3.214992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [363..460] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [461..665] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [666..900] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [901..1040] | 1.12 | Solution structure of PHD domain in death inducer-obliterator 1(DIO-1) | |
| 7 | View Details | [1041..1263] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1264..1407] | 1.23 | No description for 2dmeA was found. | |
| 9 | View Details | [1408..1558] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1559..1705] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1706..1839] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1840..2016] | 2.098994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)