













| General Information: |
|
| Name(s) found: |
Hsp70Aa-PA /
FBpp0081986
[FlyBase]
Hsp70Ab-PA / FBpp0081956 [FlyBase] |
| Description(s) found:
Found 63 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to heat
[IMP][ISS][NAS]
response to hypoxia [IEP][IMP] heat shock-mediated polytene chromosome puffing [IMP] |
| Molecular Function: |
ATP binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPAIGIDLGT TYSCVGVYQH GKVEIIANDQ GNRTTPSYVA FTDSERLIGD PAKNQVAMNP 60
61 RNTVFDAKRL IGRKYDDPKI AEDMKHWPFK VVSDGGKPKI GVEYKGESKR FAPEEISSMV 120
121 LTKMKETAEA YLGESITDAV ITVPAYFNDS QRQATKDAGH IAGLNVLRII NEPTAAALAY 180
181 GLDKNLKGER NVLIFDLGGG TFDVSILTID EGSLFEVRST AGDTHLGGED FDNRLVTHLA 240
241 DEFKRKYKKD LRSNPRALRR LRTAAERAKR TLSSSTEATI EIDALFEGQD FYTKVSRARF 300
301 EELCADLFRN TLQPVEKALN DAKMDKGQIH DIVLVGGSTR IPKVQSLLQD FFHGKNLNLS 360
361 INPDEAVAYG AAVQAAILSG DQSGKIQDVL LVDVAPLSLG IETAGGVMTK LIERNCRIPC 420
421 KQTKTFSTYA DNQPGVSIQV YEGERAMTKD NNALGTFDLS GIPPAPRGVP QIEVTFDLDA 480
481 NGILNVSAKE MSTGKAKNIT IKNDKGRLSQ AEIDRMVNEA EKYADEDEKH RQRITSRNAL 540
541 ESYVFNVKQA VEQAPAGKLD EADKNSVLDK CNDTIRWLDS NTTAEKEEFD HKLEELTRHC 600
601 SPIMTKMHQQ GAGAGAGGPG ANCGQQAGGF GGYSGPTVEE VD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
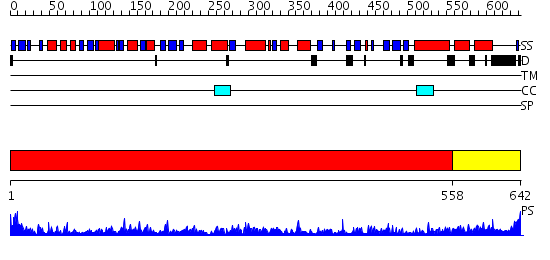
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..557] | 1000.0 | crystal structure of bovine hsc70(aa1-554)E213A/D214A mutant | |
| 2 | View Details | [558..642] | 49.69897 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)