













| General Information: |
|
| Name(s) found: |
mus309-PA /
FBpp0081910
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1487 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
reciprocal meiotic recombination
[IMP]
DNA replication [IEA] DNA duplex unwinding [IDA] double-strand break repair via homologous recombination [IMP] double-strand break repair via synthesis-dependent strand annealing [IMP] strand displacement [IDA] response to DNA damage stimulus [IGI][IMP] DNA synthesis involved in DNA repair [IMP] double-strand break repair [IMP] |
| Molecular Function: |
ATP binding
[IEA]
DNA helicase activity [ISS] DNA-dependent ATPase activity [IDA] DNA strand annealing activity [IDA] ATP-dependent 3'-5' DNA helicase activity [IDA] helicase activity [ISS] Y-form DNA binding [IDA] ATP-dependent DNA helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKKPVAQRK QLTLSSFIGL DGNSQSQPKS RAASVRSKPP AVYNPIFLDA SSSDDETTEI 60
61 SSQSNNGTIA TKKSSRDPRT AKLKKHTYLD LSVSPLAKLS AKKYARDSPK KPTSLDLSVS 120
121 PLAELLAKKS DRDSPKKPVQ NENSYTYRGL SESPVENKSI GDTLRKPPQK ERKTSIVWLS 180
181 DSPEKKVTQN ERKILDSPLQ RFSFEDFPNK ENGNRHHLLT LPDSPPPPQP VKKPEKTMWQ 240
241 NETKTIQDKD SPANPLVSNN LASISTLLDS SRAPNTYKGS SRNLFEDSPE KSGSGEQGYK 300
301 LGSAKENEIP TKPATASLER NSVTSSPSPA APLKPRYSVA FDNSLADYLK DLAQNDNFSI 360
361 DPNKQNTETL KSTLGFFRNT YVELMEKYCS LIDQIPAMHF NEIAGFQPNT FLKLKVMRQK 420
421 FKARTQLVQN SLDKKESQLK AEQEALEKEE IEMQAEQAQQ TVLSSSSPEK SRPIMPLPKV 480
481 QEIKDEKIPN RNQLIHDLCG EPDNFSPPSS PRDTQLIPKR QQLINDLCGE PDDFSPPSKQ 540
541 NDPHLLRKCE ELVHDLCEEP DDYLAQSMML DGDLEEEQLN GPTQGTTTSG MDDDEDDLEG 600
601 LLAEIEDEHQ KMQGRRSEFN GYSYKELEAV KVKEKHKETP INISLDDDGF PEYDEAMFEQ 660
661 MHSQAAANKS RVSSAGPSTS KSVVPTKQTS ALHSQKLSGN FHANVHNDGI TGEFDGQKFE 720
721 HSTRLMHGLS YSFGLKSFRP NQLQVINATL LGNDCFVLMP TGGGKSLCYQ LPAILTEGVT 780
781 IVISPLKSLI FDQINKLASL DICAKSLSGE QKMADVMAIY RDLESQPPMV KLLYVTPEKI 840
841 SSSARFQDTL DTLNSNNYIS RFVIDEAHCV SQWGHDFRPD YKKLGVLKKR FPNVPTIALT 900
901 ATATPRVRLD ILAQLNLKNC KWFLSSFNRS NLRYRVLPKK GVSTLDDISR YIRSKPQHFS 960
961 GIIYCLSRKE CDETSKKMCK DGVRAVSYHA GLTDTDRESR QKDWLTGKMR VICATVAFGM1020
1021 GIDKPDVRFV LHYSLPKSIE GYYQEAGRAG RDGDVADCIL YYNYSDMLRI KKMLDSDKAL1080
1081 QYNVKKIHVD NLYRIVGYCE NLTDCRRAQQ LDYFGEHFTS EQCLENRETA CDNCINKRAY1140
1141 KAVDALEHAR KAARAVKDLC SGRSRFTLLH IADVLKGSKI KKIIDFNHHK TPHHGVLKDW1200
1201 DKNDVHRLLR KMVIDGFLRE DLIFTNDFPQ AYLYLGNNIS KLMEGTPNFE FAVTKNAKEA1260
1261 KAAVGSVSDG ATSSTADGQS GMREIHERCY TDLLDLCRTI ASQRNVTMAS IMNIQALKSM1320
1321 AETLPITEKD MCSIPHVTKA NFDKYGAKLL EITSNYASEK LLMQAVLDEE EEQAAAKQRP1380
1381 STSGWNNESV DWDMAVASQG NANTSGASGF NSFRAGKRKK IYKSGASKRY KTSTTSPAAR1440
1441 KTTSARGRGG RAGAKRAESS ASSASGWKSK KTGNSFGFDL MPLPGSK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
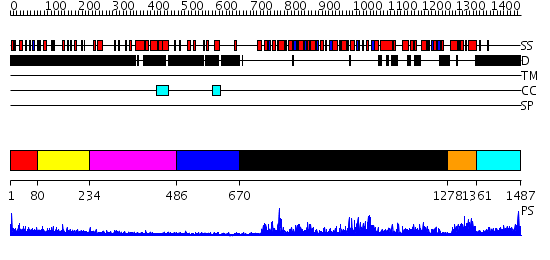
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [80..233] | 1.252988 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [234..485] | 1.263996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [486..669] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [670..1277] | 121.0 | No description for 2v1xA was found. | |
| 6 | View Details | [1278..1360] | 22.39794 | E. coli RecQ HRDC domain | |
| 7 | View Details | [1361..1487] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|