













| General Information: |
|
| Name(s) found: |
MTA1-like-PB /
FBpp0078383
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 844 amino acids |
Gene Ontology: |
|
| Cellular Component: |
histone deacetylase complex
[IDA]
|
| Biological Process: |
phagocytosis, engulfment
[IMP]
regulation of transcription, DNA-dependent [IEA] |
| Molecular Function: |
transcription factor activity
[IEA]
zinc ion binding [IEA] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATNMYRVGD YVYVETTPNS PYLIRRIEEL NKNQTGNVEA KVMCFYRRRD LPNPLVQLAD 60
61 KHQPALDEER TSPTQTSGGG GSATGNSGSN SSGTSNNNSS STAIGGGAGG SGGAGGDAEK 120
121 GEALTSKQRY QIKHRELFLS RQVESIPATQ IRGKCSVTLL NETESLQSYL NKDDTFFYCL 180
181 VFDPNQKTLL ADKGEIRVGS RYQCDIPAKL KDTATDDRKL EELESLVWTP EHSLTDRKID 240
241 QFLVVSRSIG TFARALDCSS SVKQPSLHMS AAAASRDITL FHAMNILHKH EYSIEESMSS 300
301 LVPSTGPVLC RDEIEDWSAS EANLFEEALE KYGKDFNDIR QDFLPWKTLK QIIEYYYMWK 360
361 TTDRYVQQKR VKAVEAELKL KQVYIPQYNN NGKGNGTSTK AGGGIYNGTT NGSTDLSSNG 420
421 KPCESCGTTK SSQWNSVSSG HSTSRLCLSC WEYWRRYGSM KSATKGDAGE GDAKKKSSSA 480
481 ASTPTATLAG LATTPTAVVD LNDDEKISDL TNRQLHRCSI VNCGKEFKLK THLARHYAQA 540
541 HGIAISSGSP RPIMKTRTAF YLHTNPMTRV ARAICRSIVK PKKAARQSAY AINAMLVKQE 600
601 FTNRISGKSQ AEIKKLLLLK PKDRGSVTKI ANRLGAPGSG PHEWLVLTPK DKMPLPAVVS 660
661 FPKPPKAPDG SLVYDRVPNK SPDVVAVPAD KELTIIPTQA TSTIRKRAHE DQQLNGTEVT 720
721 IVPSGPPAKR PNKDPMPSHC PSPEQFAAMM AASGQPLSRH HLNGKQKIAQ MARGGNGRKQ 780
781 VISWMDAPDD VYFRANDTHK KTRKILSAVD LRRAARKPWR TLPIKPAAPE PSSRPIESQI 840
841 VILD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
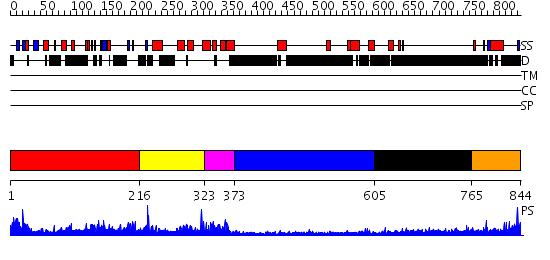
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | 2.72 | Crystal structure of the proximal BAH domain of polybromo | |
| 2 | View Details | [216..322] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [323..372] | 19.221849 | Solution structure of the myb-like DNA-binding domain of mouse MTA3 protein | |
| 4 | View Details | [373..604] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [605..764] | 3.304996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [765..844] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)