













| General Information: |
|
| Name(s) found: |
FBpp0310716
[FlyBase]
FBpp0306391 [FlyBase] |
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 478 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to hypoxia
[IMP]
protein localization [IMP] oxidation reduction [IEA] response to DNA damage stimulus [IMP] open tracheal system development [NAS] |
| Molecular Function: |
L-ascorbic acid binding
[IEA]
zinc ion binding [IEA] oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen [IEA] iron ion binding [IEA] peptidyl-proline 4-dioxygenase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRGRGKVRD SASHSGSHSA SKSAMDPPRC SICGTQQQLL RCAKCKAVYY CSPAHQHLHW 60
61 PDHRTECRLL TRQKLNSSNN NKQQQRQQIQ QLQQAVASAN LECSGAGANC STAQMMTPAH 120
121 QAQSWPAEVD NLLNLLGQPG SQEKAAAAET ETGQRQQQHQ HHHHNGEKSS SYQIGLADAS 180
181 FMGSGSERRY EDLCRNIISD MNQYGLSVVD DFLGMETGLK ILNEVRSMYN AGAFQDGQVV 240
241 TNQTPDAPAV RGDKIRGDKI KWVGGNEPGC SNVWYLTNQI DSVVYRVNTM KDNGILGNYH 300
301 IRERTRAMVA CYPGSGTHYV MHVDNPQKDG RVITAIYYLN INWDARESGG ILRIRPTPGT 360
361 TVADIEPKFD RLIFFWSDIR NPHEVQPAHR TRYAITVWYF DAKEREEALI RAKLENSKTN 420
421 NLAAQAQAQQ AEPDSTTTPP AAPASSASSL PVSMSTGTGA LNANVSSNSC ATSSEICT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
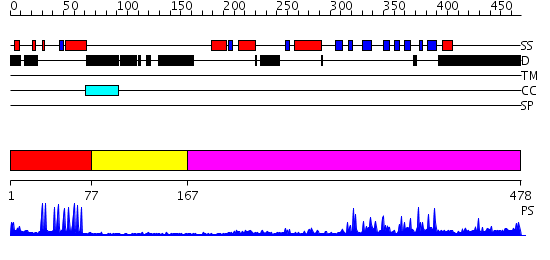
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | 10.30103 | No description for 2d8qA was found. | |
| 2 | View Details | [77..166] | 1.01 | No description for 2imlA was found. | |
| 3 | View Details | [167..478] | 59.045757 | No description for 2hbtA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)