













| General Information: |
|
| Name(s) found: |
abs-PA /
FBpp0078606
[FlyBase]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 619 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
catalytic step 2 spliceosome [IDA] precatalytic spliceosome [IDA] |
| Biological Process: |
apoptosis
[NAS]
nervous system development [NAS] establishment of cell polarity [NAS] anatomical structure morphogenesis [NAS] nuclear mRNA splicing, via spliceosome [IC] |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] zinc ion binding [IEA] ATP-dependent helicase activity [ISS] helicase activity [ISS] ATP-dependent RNA helicase activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAHVKRYRRS SKSSEEGDLD NEDYVPYVPV KERKKQHMIK LGRIVQLVSE TAQPKSSSEN 60
61 ENEDDSQGAH DVETWGRKYN ISLLDQHTEL KKIAEAKKLS AVEKQLREEE KIMESIAQQK 120
121 ALMGVAELAK GIQYEQPIKT AWKPPRYIRE MSEEEREAVR HELRILVEGE TPSPPIRSFR 180
181 EMKFPKGILN GLAAKGIKNP TPIQVQGLPT VLAGRDLIGI AFTGSGKTLV FVLPVIMFAL 240
241 EQEYSLPFER NEGPYGLIIC PSRELAKQTH EIIQHYSKHL QACGMPEIRS CLAMGGLPVS 300
301 EALDVISRGV HIVVATPGRL MDMLDKKILT LDMCRYLCMD EADRMIDMGF EEDVRTIFSF 360
361 FKGQRQTLLF SATMPKKIQN FARSALVKPV TINVGRAGAA SMNVTQQVEY VKQEAKVVYL 420
421 LDCLQKTAPP VLIFAEKKQD VDCIHEYLLL KGVEAVAIHG GKDQEERSRA VDAYRVGKKD 480
481 VLVATDVASK GLDFPNVQHV INYDMPDDIE NYVHRIGRTG RSNTKGLATT LINKTTEQSV 540
541 LLDLKHLLIE GKQEVPDFLD ELAPETEHQH LDLGDSHGCT YCGGLGHRIT ECPKLEAVQN 600
601 KQASNIGRRD YLSNTAADY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
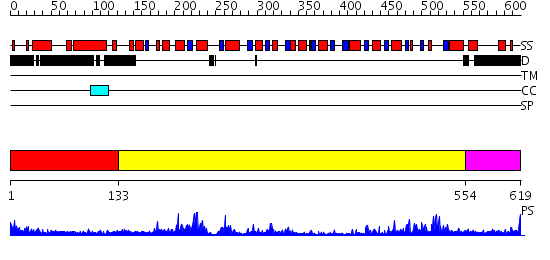
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [133..553] | 102.0 | No description for 2db3A was found. | |
| 3 | View Details | [554..619] | 19.522879 | No description for 2d7dA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)